Cyclic modular beta-sheets
- PMID: 17295482
- PMCID: PMC2597679
- DOI: 10.1021/ja0667965
Cyclic modular beta-sheets
Abstract
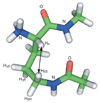
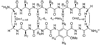
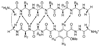
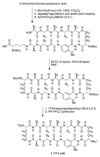
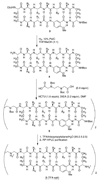
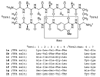
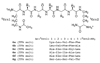
The development of peptide beta-hairpins is problematic, because folding depends on the amino acid sequence and changes to the sequence can significantly decrease folding. Robust beta-hairpins that can tolerate such changes are attractive tools for studying interactions involving protein beta-sheets and developing inhibitors of these interactions. This paper introduces a new class of peptide models of protein beta-sheets that addresses the problem of separating folding from the sequence. These model beta-sheets are macrocyclic peptides that fold in water to present a pentapeptide beta-strand along one edge; the other edge contains the tripeptide beta-strand mimic Hao [JACS 2000, 122, 7654] and two additional amino acids. The pentapeptide and Hao-containing peptide strands are connected by two delta-linked ornithine (deltaOrn) turns [JACS 2003, 125, 876]. Each deltaOrn turn contains a free alpha-amino group that permits the linking of individual modules to form divalent beta-sheets. These "cyclic modular beta-sheets" are synthesized by standard solid-phase peptide synthesis of a linear precursor followed by solution-phase cyclization. Eight cyclic modular beta-sheets 1a-1h containing sequences based on beta-amyloid and macrophage inflammatory protein 2 were synthesized and characterized by 1H NMR. Linked cyclic modular beta-sheet 2, which contains two modules of 1b, was also synthesized and characterized. 1H NMR studies show downfield alpha-proton chemical shifts, deltaOrn delta-proton magnetic anisotropy, and NOE cross-peaks that establish all compounds but 1c and 1g to be moderately or well folded into a conformation that resembles a beta-sheet. Pulsed-field gradient NMR diffusion experiments show little or no self-association at low (</=2 mM) concentrations. Changes to the residues in the Hao-containing strands of 1c and 1g improve folding and show that folding of the structures can be enhanced without altering the sequence of the pentapeptide strand. Well-folded cyclic modular beta-sheets 1a, 1b, and 1f each have a phenylalanine directly across from Hao, suggesting that cyclic modular beta-sheets containing aromatic residues across from Hao are better folded.
Figures








References
-
- Loughlin WA, Tyndall JDA, Glenn MP, Fairlie DP. Chem. Rev. 2004;104:6085–6117. - PubMed
-
- DeLano WL, Ultsch MH, de Vos AM, Wells JA. Science. 2000;287:1279–1283. - PubMed
- Fasan R, Dias RLA, Moehle K, Zerbe O, Vrijbloed JW, Obrecht D, Robinson JA. Angew. Chem. Int. Ed. 2004;43:2109–2112. - PubMed
- Butterfield SM, Cooper WJ, Waters ML. J. Am. Chem. Soc. 2005;127:24–25. - PubMed
- Dias RLA, Fasan R, Moehle K, Renard A, Obrecht D, Robinson JA. J. Am. Chem. Soc. 2006;128:2726–2732. - PubMed
-
- Smith CK, Regan L. Acc. Chem. Res. 1997;30:153–161.
- Blanco F, Ramírez-Alvarado M, Serrano L. Curr. Opin. Struct. Biol. 1998;8:107–111. - PubMed
- Gellman SH. Curr. Opin. Chem. Biol. 1998;2:717–725. - PubMed
- Lacroix E, Kortemme T, de la Paz ML, Serrano L. Curr. Opin. Struct. Biol. 1999;9:487–493. - PubMed
- Ramírez-Alvarado M, Kortemme T, Blanco FJ, Serrano L. Bioorg. Med. Chem. 1999;7:93–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

