Elimination of laboratory ozone leads to a dramatic improvement in the reproducibility of microarray gene expression measurements
- PMID: 17295919
- PMCID: PMC1800842
- DOI: 10.1186/1472-6750-7-8
Elimination of laboratory ozone leads to a dramatic improvement in the reproducibility of microarray gene expression measurements
Abstract
Background: Environmental ozone can rapidly degrade cyanine 5 (Cy5), a fluorescent dye commonly used in microarray gene expression studies. Cyanine 3 (Cy3) is much less affected by atmospheric ozone. Degradation of the Cy5 signal relative to the Cy3 signal in 2-color microarrays will adversely reduce the Cy5/Cy3 ratio resulting in unreliable microarray data.
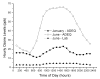
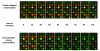
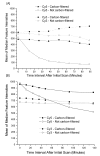
Results: Ozone in central Arkansas typically ranges between approximately 22 ppb to approximately 46 ppb and can be as high as 60-100 ppb depending upon season, meteorological conditions, and time of day. These levels of ozone are common in many areas of the country during the summer. A carbon filter was installed in the laboratory air handling system to reduce ozone levels in the microarray laboratory. In addition, the airflow was balanced to prevent non-filtered air from entering the laboratory. These modifications reduced the ozone within the microarray laboratory to approximately 2-4 ppb. Data presented here document reductions in Cy5 signal on both in-house produced microarrays and commercial microarrays as a result of exposure to unfiltered air. Comparisons of identically hybridized microarrays exposed to either carbon-filtered or unfiltered air demonstrated the protective effect of carbon-filtration on microarray data as indicated by Cy5 and Cy3 intensities. LOWESS normalization of the data was not able to completely overcome the effect of ozone-induced reduction of Cy5 signal. Experiments were also conducted to examine the effects of high humidity on microarray quality. Modest, but significant, increases in Cy5 and Cy3 signal intensities were observed after 2 or 4 hours at 98-99% humidity compared to 42% humidity.
Conclusion: Simple installation of carbon filters in the laboratory air handling system resulted in low and consistent ozone levels. This allowed the accurate determination of gene expression by microarray using Cy5 and Cy3 fluorescent dyes.
Figures
References
-
- U.S. EPA http://www.epa.gov/epahome/ozone.htm
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

