Regulation of rRNA synthesis by TATA-binding protein-associated factor Mot1
- PMID: 17296733
- PMCID: PMC1899949
- DOI: 10.1128/MCB.00054-07
Regulation of rRNA synthesis by TATA-binding protein-associated factor Mot1
Abstract
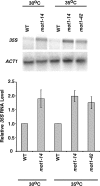
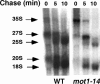
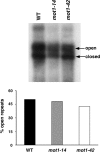
Mot1 is an essential, conserved, TATA-binding protein (TBP)-associated factor in Saccharomyces cerevisiae with well-established roles in the global control of RNA polymerase II (Pol II) transcription. Previous results have suggested that Mot1 functions exclusively in Pol II transcription, but here we report a novel role for Mot1 in regulating transcription by RNA polymerase I (Pol I). In vivo, Mot1 is associated with the ribosomal DNA, and loss of Mot1 results in decreased rRNA synthesis. Consistent with a direct role for Mot1 in Pol I transcription, Mot1 also associates with the Pol I promoter in vitro in a reaction that depends on components of the Pol I general transcription machinery. Remarkably, in addition to Mot1's role in initiation, rRNA processing is delayed in mot1 cells. Taken together, these results support a model in which Mot1 affects the rate and efficiency of rRNA synthesis by both direct and indirect mechanisms, with resulting effects on transcription activation and the coupling of rRNA synthesis to processing.
Figures


Similar articles
-
TATA-binding protein variants that bypass the requirement for Mot1 in vivo.J Biol Chem. 2009 Feb 13;284(7):4525-35. doi: 10.1074/jbc.M808951200. Epub 2008 Dec 21. J Biol Chem. 2009. PMID: 19098311 Free PMC article.
-
Mot1-mediated control of transcription complex assembly and activity.EMBO J. 2005 May 4;24(9):1717-29. doi: 10.1038/sj.emboj.7600646. Epub 2005 Apr 7. EMBO J. 2005. PMID: 15861138 Free PMC article.
-
Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae.Mol Cell Biol. 2002 Dec;22(23):8122-34. doi: 10.1128/MCB.22.23.8122-8134.2002. Mol Cell Biol. 2002. PMID: 12417716 Free PMC article.
-
One small step for Mot1; one giant leap for other Swi2/Snf2 enzymes?Biochim Biophys Acta. 2011 Sep;1809(9):488-96. doi: 10.1016/j.bbagrm.2011.05.012. Epub 2011 May 30. Biochim Biophys Acta. 2011. PMID: 21658482 Free PMC article. Review.
-
Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing.Curr Opin Cell Biol. 2005 Jun;17(3):281-6. doi: 10.1016/j.ceb.2005.04.001. Curr Opin Cell Biol. 2005. PMID: 15901498 Review.
Cited by
-
Psoralen Crosslinking-Chromatin Endogenous Cleavage Assay to Examine Histone DNA Interactions of Active and Inactive rRNA Genes.Methods Mol Biol. 2025;2919:133-154. doi: 10.1007/978-1-0716-4486-7_8. Methods Mol Biol. 2025. PMID: 40257561
-
TATA-binding protein variants that bypass the requirement for Mot1 in vivo.J Biol Chem. 2009 Feb 13;284(7):4525-35. doi: 10.1074/jbc.M808951200. Epub 2008 Dec 21. J Biol Chem. 2009. PMID: 19098311 Free PMC article.
-
Conserved regulators of nucleolar size revealed by global phenotypic analyses.Sci Signal. 2013 Aug 20;6(289):ra70. doi: 10.1126/scisignal.2004145. Sci Signal. 2013. PMID: 23962978 Free PMC article.
-
Antibiotic-Induced Treatments Reveal Stress-Responsive Gene Expression in the Endangered Lichen Lobaria pulmonaria.J Fungi (Basel). 2022 Jun 12;8(6):625. doi: 10.3390/jof8060625. J Fungi (Basel). 2022. PMID: 35736108 Free PMC article.
-
Rpd3- and spt16-mediated nucleosome assembly and transcriptional regulation on yeast ribosomal DNA genes.Mol Cell Biol. 2013 Jul;33(14):2748-59. doi: 10.1128/MCB.00112-13. Epub 2013 May 20. Mol Cell Biol. 2013. PMID: 23689130 Free PMC article.
References
-
- Angermayr, M., U. Oechsner, and W. Bandlow. 2003. Reb1p-dependent DNA bending effects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J. Biol. Chem. 278:17918-17926. - PubMed
-
- Auble, D. T., K. E. Hansen, C. G. F. Mueller, W. S. Lane, J. Thorner, and S. Hahn. 1994. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8:1920-1934. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
