DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur
- PMID: 17296795
- PMCID: PMC2063980
- DOI: 10.1083/jcb.200609014
DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur
Abstract
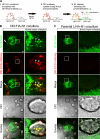
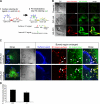
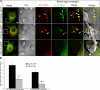
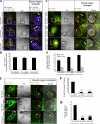
Cleavage of Notch by furin is required to generate a mature, cell surface heterodimeric receptor that can be proteolytically activated to release its intracellular domain, which functions in signal transduction. Current models propose that ligand binding to heterodimeric Notch (hNotch) induces a disintegrin and metalloprotease (ADAM) proteolytic release of the Notch extracellular domain (NECD), which is subsequently shed and/or endocytosed by DSL ligand cells. We provide evidence for NECD release and internalization by DSL ligand cells, which, surprisingly, did not require ADAM activity. However, losses in either hNotch formation or ligand endocytosis significantly decreased NECD transfer to DSL ligand cells, as well as signaling in Notch cells. Because endocytosis-defective ligands bind hNotch, but do not dissociate it, additional forces beyond those produced through ligand binding must function to disrupt the intramolecular interactions that keep hNotch intact and inactive. Based on our findings, we propose that mechanical forces generated during DSL ligand endocytosis function to physically dissociate hNotch, and that dissociation is a necessary step in Notch activation.
Figures



Similar articles
-
Microfibrillar proteins MAGP-1 and MAGP-2 induce Notch1 extracellular domain dissociation and receptor activation.J Biol Chem. 2006 Apr 14;281(15):10089-97. doi: 10.1074/jbc.M600298200. Epub 2006 Feb 20. J Biol Chem. 2006. PMID: 16492672
-
Endocytosis-independent mechanisms of Delta ligand proteolysis.Exp Cell Res. 2006 May 1;312(8):1345-60. doi: 10.1016/j.yexcr.2005.12.037. Epub 2006 Feb 17. Exp Cell Res. 2006. PMID: 16487968
-
Notch1 endocytosis is induced by ligand and is required for signal transduction.Biochim Biophys Acta. 2016 Jan;1863(1):166-77. doi: 10.1016/j.bbamcr.2015.10.021. Epub 2015 Oct 30. Biochim Biophys Acta. 2016. PMID: 26522918
-
RIPping notch apart: a new role for endocytosis in signal transduction?Sci STKE. 2000 Apr 25;2000(29):pe1. doi: 10.1126/stke.2000.29.pe1. Sci STKE. 2000. PMID: 11752592 Review.
-
The role of ligand endocytosis in notch signalling.Biol Cell. 2021 Oct;113(10):401-418. doi: 10.1111/boc.202100009. Epub 2021 Jun 4. Biol Cell. 2021. PMID: 34038572 Review.
Cited by
-
Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch.Dev Cell. 2015 Jun 22;33(6):729-36. doi: 10.1016/j.devcel.2015.05.004. Epub 2015 Jun 4. Dev Cell. 2015. PMID: 26051539 Free PMC article.
-
Endocytosis and control of Notch signaling.Curr Opin Cell Biol. 2012 Aug;24(4):534-40. doi: 10.1016/j.ceb.2012.06.006. Epub 2012 Jul 18. Curr Opin Cell Biol. 2012. PMID: 22818956 Free PMC article. Review.
-
The Bro1-domain protein, EGO-2, promotes Notch signaling in Caenorhabditis elegans.Genetics. 2007 Aug;176(4):2265-77. doi: 10.1534/genetics.107.071225. Epub 2007 Jul 1. Genetics. 2007. PMID: 17603118 Free PMC article.
-
Enhancement of Notch receptor maturation and signaling sensitivity by Cripto-1.J Cell Biol. 2009 Nov 2;187(3):343-53. doi: 10.1083/jcb.200905105. J Cell Biol. 2009. PMID: 19948478 Free PMC article.
-
S/T phosphorylation of DLL1 is required for full ligand activity in vitro but dispensable for DLL1 function in vivo during embryonic patterning and marginal zone B cell development.Mol Cell Biol. 2014 Apr;34(7):1221-33. doi: 10.1128/MCB.00965-13. Epub 2014 Jan 21. Mol Cell Biol. 2014. PMID: 24449764 Free PMC article.
References
-
- Blaumueller, C.M., H. Qi, P. Zagouras, and S. Artavanis-Tsakonas. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 90:281–291. - PubMed
-
- Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail Jr., A. Doedens, P. Cumano, B.R.A. Roux, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the Disintegrin-Metalloprotease TACE. Mol. Cell. 5:207–216. - PubMed
-
- Brown, M.S., J. Ye, R.B. Rawson, and J.L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 100:391–398. - PubMed
-
- Bush, G., G. diSibio, A. Miyamoto, J.B. Denault, R. Leduc, and G. Weinmaster. 2001. Ligand-induced signaling in the absence of furin processing of Notch1. Dev. Biol. 229:494–502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

