Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation
- PMID: 17296936
- PMCID: PMC1796997
- DOI: 10.1073/pnas.0608056104
Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation
Abstract
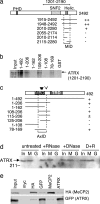
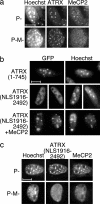
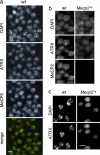
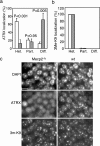
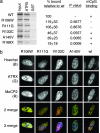
Mutations in the human methyl-CpG-binding protein gene MECP2 cause the neurological disorder Rett syndrome and some cases of X-linked mental retardation (XLMR). We report that MeCP2 interacts with ATRX, a SWI2/SNF2 DNA helicase/ATPase that is mutated in ATRX syndrome (alpha-thalassemia/mental retardation, X-linked). MeCP2 can recruit the helicase domain of ATRX to heterochromatic foci in living mouse cells in a DNA methylation-dependent manner. Also, ATRX localization is disrupted in neurons of Mecp2-null mice. Point mutations within the methylated DNA-binding domain of MeCP2 that cause Rett syndrome or X-linked mental retardation inhibit its interaction with ATRX in vitro and its localization in vivo without affecting methyl-CpG binding. We propose that disruption of the MeCP2-ATRX interaction leads to pathological changes that contribute to mental retardation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Hong EJ, West AE, Greenberg ME. Curr Opin Neurobiol. 2005;15:21–28. - PubMed
-
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Nat Genet. 1998;19:187–191. - PubMed
-
- Nan X, Campoy FJ, Bird A. Cell. 1997;88:471–481. - PubMed
-
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nature. 1998;393:386–389. - PubMed
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Nat Genet. 1999;23:185–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

