N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis
- PMID: 17298082
- PMCID: PMC2569195
- DOI: 10.1021/bi602436g
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis
Abstract
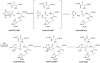
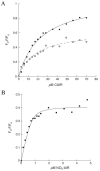
N5-Carboxyaminoimidazole ribonucleotide mutase (N5-CAIR mutase or PurE) from Escherichia coli catalyzes the reversible interconversion of N5-CAIR to carboxyaminoimidazole ribonucleotide (CAIR) with direct CO2 transfer. Site-directed mutagenesis, a pH-rate profile, DFT calculations, and X-ray crystallography together provide new insight into the mechanism of this unusual transformation. These studies suggest that a conserved, protonated histidine (His45) plays an essential role in catalysis. The importance of proton transfers is supported by DFT calculations on CAIR and N5-CAIR analogues in which the ribose 5'-phosphate is replaced with a methyl group. The calculations suggest that the nonaromatic tautomer of CAIR (isoCAIR) is only 3.1 kcal/mol higher in energy than its aromatic counterpart, implicating this species as a potential intermediate in the PurE-catalyzed reaction. A structure of wild-type PurE cocrystallized with 4-nitroaminoimidazole ribonucleotide (NO2-AIR, a CAIR analogue) and structures of H45N and H45Q PurEs soaked with CAIR have been determined and provide the first insight into the binding of an intact PurE substrate. A comparison of 19 available structures of PurE and PurE mutants in apo and nucleotide-bound forms reveals a common, buried carboxylate or CO2 binding site for CAIR and N5-CAIR in a hydrophobic pocket in which the carboxylate or CO2 interacts with backbone amides. This work has led to a mechanistic proposal in which the carboxylate orients the substrate for proton transfer from His45 to N5-CAIR to form an enzyme-bound aminoimidazole ribonucleotide (AIR) and CO2 intermediate. Subsequent movement of the aminoimidazole moiety of AIR reorients it for addition of CO2 at C4 to generate isoCAIR. His45 is now in a position to remove a C4 proton to produce CAIR.
Figures

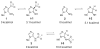



Similar articles
-
Biochemical and structural studies of N5-carboxyaminoimidazole ribonucleotide mutase from the acidophilic bacterium Acetobacter aceti.Biochemistry. 2006 Jul 11;45(27):8193-208. doi: 10.1021/bi060465n. Biochemistry. 2006. PMID: 16819818
-
Evidence for the direct transfer of the carboxylate of N5-carboxyaminoimidazole ribonucleotide (N5-CAIR) to generate 4-carboxy-5-aminoimidazole ribonucleotide catalyzed by Escherichia coli PurE, an N5-CAIR mutase.Biochemistry. 1999 Mar 9;38(10):3012-8. doi: 10.1021/bi9827159. Biochemistry. 1999. PMID: 10074353
-
Exploration of N5-CAIR Mutase Novel Inhibitors from Pyrococcus horikoshii OT3: A Computational Study.J Comput Biol. 2019 May;26(5):457-472. doi: 10.1089/cmb.2018.0248. Epub 2019 Feb 19. J Comput Biol. 2019. PMID: 30785305
-
When the host senses the microbiota's metabolism - the interplay between food and microbiota in C. elegans.FEBS J. 2025 Jun;292(11):2767-2770. doi: 10.1111/febs.70054. Epub 2025 Mar 9. FEBS J. 2025. PMID: 40059294 Free PMC article. Review.
-
Orotidine 5'-Monophosphate Decarboxylase: Probing the Limits of the Possible for Enzyme Catalysis.Acc Chem Res. 2018 Apr 17;51(4):960-969. doi: 10.1021/acs.accounts.8b00059. Epub 2018 Mar 29. Acc Chem Res. 2018. PMID: 29595949 Free PMC article. Review.
Cited by
-
The Recognition of Identical Ligands by Unrelated Proteins.ACS Chem Biol. 2015 Dec 18;10(12):2772-84. doi: 10.1021/acschembio.5b00683. Epub 2015 Oct 12. ACS Chem Biol. 2015. PMID: 26421501 Free PMC article.
-
Bacterial purine metabolism modulates C. elegans development and stress tolerance via DAF-16.FEBS J. 2025 Jun;292(11):2771-2783. doi: 10.1111/febs.17363. Epub 2024 Dec 21. FEBS J. 2025. PMID: 39708289 Free PMC article.
-
Probing the active center of benzaldehyde lyase with substitutions and the pseudosubstrate analogue benzoylphosphonic acid methyl ester.Biochemistry. 2008 Jul 22;47(29):7734-43. doi: 10.1021/bi8004413. Epub 2008 Jun 21. Biochemistry. 2008. PMID: 18570438 Free PMC article.
-
Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.Acta Crystallogr D Biol Crystallogr. 2011 Aug;67(Pt 8):707-15. doi: 10.1107/S0907444911023821. Epub 2011 Jul 12. Acta Crystallogr D Biol Crystallogr. 2011. PMID: 21795812 Free PMC article.
-
The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.Proc Natl Acad Sci U S A. 2021 Sep 28;118(39):e2106202118. doi: 10.1073/pnas.2106202118. Proc Natl Acad Sci U S A. 2021. PMID: 34548397 Free PMC article.
References
-
- Mueller EJ, Meyer E, Rudolph J, Davisson VJ, Stubbe J. N5-carboxyaminoimidazole ribonucleotide: evidence for a new intermediate and two new enzymatic activities in the de novo purine biosynthetic pathway of Escherichia coli. Biochemistry. 1994;33:2269–78. - PubMed
-
- Firestine SM, Davisson VJ. Carboxylases in de novo purine biosynthesis. Characterization of the Gallus gallus bifunctional enzyme. Biochemistry. 1994;33:11917–26. - PubMed
-
- Mathews II, Kappock TJ, Stubbe J, Ealick SE. Crystal structure of Escherichia coli PurE, an unusual mutase in the purine biosynthetic pathway. Structure Fold Des. 1999;7:1395–406. - PubMed
-
- Firestine SM, Poon SW, Mueller EJ, Stubbe J, Davisson VJ. Reactions catalyzed by 5-aminoimidazole ribonucleotide carboxylases from Escherichia coli and Gallus gallus: a case for divergent catalytic mechanisms. Biochemistry. 1994;33:11927–34. - PubMed
-
- Litchfield GJ, Shaw G. Purines, Pyrimidines, and Imidazoles. Part XXXVIII. A Kinetic Study of the Decarboxylation of 5-Amino-1-β-D-ribofuranosylimidazole-4-carboxylic Acid 5’-Phosphate and Related Compounds. J Chem Soc (B) 1971:1474–1484.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

