System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock
- PMID: 17298173
- PMCID: PMC1783671
- DOI: 10.1371/journal.pbio.0050034
System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock
Abstract
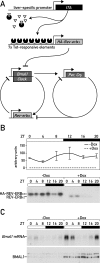
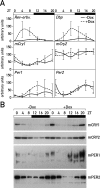
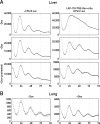
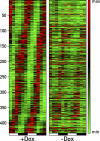
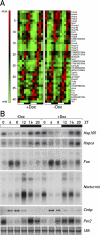
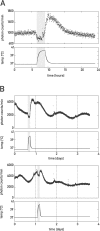
The mammalian circadian timing system consists of a master pacemaker in neurons of the suprachiasmatic nucleus (SCN) and clocks of a similar molecular makeup in most peripheral body cells. Peripheral oscillators are self-sustained and cell autonomous, but they have to be synchronized by the SCN to ensure phase coherence within the organism. In principle, the rhythmic expression of genes in peripheral organs could thus be driven not only by local oscillators, but also by circadian systemic signals. To discriminate between these mechanisms, we engineered a mouse strain with a conditionally active liver clock, in which REV-ERBalpha represses the transcription of the essential core clock gene Bmal1 in a doxycycline-dependent manner. We examined circadian liver gene expression genome-wide in mice in which hepatocyte oscillators were either running or arrested, and found that the rhythmic transcription of most genes depended on functional hepatocyte clocks. However, we discovered 31 genes, including the core clock gene mPer2, whose expression oscillated robustly irrespective of whether the liver clock was running or not. By contrast, in liver explants cultured in vitro, circadian cycles of mPer2::luciferase bioluminescence could only be observed when hepatocyte oscillators were operational. Hence, the circadian cycles observed in the liver of intact animals without functional hepatocyte oscillators were likely generated by systemic signals. The finding that rhythmic mPer2 expression can be driven by both systemic cues and local oscillators suggests a plausible mechanism for the phase entrainment of subsidiary clocks in peripheral organs.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

Comment in
-
Central and peripheral signals set the circadian liver clock.PLoS Biol. 2007 Feb;5(2):e50. doi: 10.1371/journal.pbio.0050050. Epub 2007 Jan 30. PLoS Biol. 2007. PMID: 20076659 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

