Genome-wide profiling and analysis of Arabidopsis siRNAs
- PMID: 17298187
- PMCID: PMC1820830
- DOI: 10.1371/journal.pbio.0050057
Genome-wide profiling and analysis of Arabidopsis siRNAs
Abstract
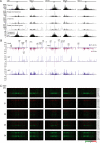
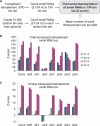
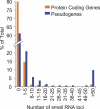
Eukaryotes contain a diversified set of small RNA-guided pathways that control genes, repeated sequences, and viruses at the transcriptional and posttranscriptional levels. Genome-wide profiles and analyses of small RNAs, particularly the large class of 24-nucleotide (nt) short interfering RNAs (siRNAs), were done for wild-type Arabidopsis thaliana and silencing pathway mutants with defects in three RNA-dependent RNA polymerase (RDR) and four Dicer-like (DCL) genes. The profiling involved direct analysis using a multiplexed, parallel-sequencing strategy. Small RNA-generating loci, especially those producing predominantly 24-nt siRNAs, were found to be highly correlated with repetitive elements across the genome. These were found to be largely RDR2- and DCL3-dependent, although alternative DCL activities were detected on a widespread level in the absence of DCL3. In contrast, no evidence for RDR2-alternative activities was detected. Analysis of RDR2- and DCL3-dependent small RNA accumulation patterns in and around protein-coding genes revealed that upstream gene regulatory sequences systematically lack siRNA-generating activities. Further, expression profiling suggested that relatively few genes, proximal to abundant 24-nt siRNAs, are regulated directly by RDR2- and DCL3-dependent silencing. We conclude that the widespread accumulation patterns for RDR2- and DCL3-dependent siRNAs throughout the Arabidopsis genome largely reflect mechanisms to silence highly repeated sequences.
Conflict of interest statement
Figures
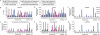

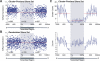
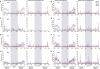
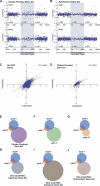
References
-
- Almeida R, Allshire RC. RNA silencing and genome regulation. Trends Cell Biol. 2005;15:251–258. - PubMed
-
- Tomari Y, Zamore PD. Perspective: Machines for RNAi. Genes Dev. 2005;19:517–529. - PubMed
-
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. - PubMed
-
- Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. - PubMed
-
- Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

