Characterization of relationships between transcriptional units and operon structures in Bacillus subtilis and Escherichia coli
- PMID: 17298663
- PMCID: PMC1808063
- DOI: 10.1186/1471-2164-8-48
Characterization of relationships between transcriptional units and operon structures in Bacillus subtilis and Escherichia coli
Abstract
Background: Operon structures play an important role in transcriptional regulation in prokaryotes. However, there have been fewer studies on complicated operon structures in which the transcriptional units vary with changing environmental conditions. Information about such complicated operons is helpful for predicting and analyzing operon structures, as well as understanding gene functions and transcriptional regulation.
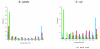
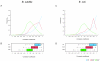
Results: We systematically analyzed the experimentally verified transcriptional units (TUs) in Bacillus subtilis and Escherichia coli obtained from ODB and RegulonDB. To understand the relationships between TUs and operons, we defined a new classification system for adjacent gene pairs, divided into three groups according to the level of gene co-regulation: operon pairs (OP) belong to the same TU, sub-operon pairs (SOP) that are at the transcriptional boundaries within an operon, and non-operon pairs (NOP) belonging to different operons. Consequently, we found that the levels of gene co-regulation was correlated to intergenic distances and gene expression levels. Additional analysis revealed that they were also correlated to the levels of conservation across about 200 prokaryotic genomes. Most interestingly, we found that functional associations in SOPs were more observed in the environmental and genetic information processes.
Conclusion: Complicated operon structures were correlated with genome organization and gene expression profiles. Such intricately regulated operons allow functional differences depending on environmental conditions. These regulatory mechanisms are helpful in accommodating the variety of changes that happen around the cell. In addition, such differences may play an important role in the evolution of gene order across genomes.
Figures
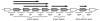



References
-
- Craven M, Page D, Shavlik J, Bockhorst J, Glasner J. A probabilistic learning approach to whole-genome operon prediction. Proc Int Conf Intell Syst Mol Biol. 2000;8:116–127. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

