Long-term trends in evolution of indels in protein sequences
- PMID: 17298668
- PMCID: PMC1805498
- DOI: 10.1186/1471-2148-7-19
Long-term trends in evolution of indels in protein sequences
Abstract
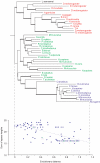
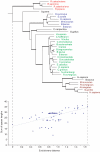
Background: In this paper we describe an analysis of the size evolution of both protein domains and their indels, as inferred by changing sizes of whole domains or individual unaligned regions or "spacers". We studied relatively early evolutionary events and focused on protein domains which are conserved among various taxonomy groups.
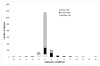
Results: We found that more than one third of all domains have a statistically significant tendency to increase/decrease in size in evolution as judged from the overall domain size distribution as well as from the size distribution of individual spacers. Moreover, the fraction of domains and individual spacers increasing in size is almost twofold larger than the fraction decreasing in size.
Conclusion: We showed that the tolerance to insertion and deletion events depends on the domain's taxonomy span. Eukaryotic domains are depleted in insertions compared to the overall test set, namely, the number of spacers increasing in size is about the same as the number of spacers decreasing in size. On the other hand, ancient domain families show some bias towards insertions or spacers which grow in size in evolution. Domains from several Gene Ontology categories also demonstrate certain tendencies for insertion or deletion events as inferred from the analysis of spacer sizes.
Figures
References
-
- Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1284. - PubMed
-
- Kummerfeld SK, Teichmann SA. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21:25–30. - PubMed
-
- Zhang J. Protein-length distributions for the three domains of life. Trends Genet. 2000;16:107–109. - PubMed
-
- Liang P, Riley M. A comparative genomics approach for studying ancestral proteins and evolution. Adv Appl Microbiol. 2001;50:39–72. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources