A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures
- PMID: 17299036
- PMCID: PMC1815236
- DOI: 10.1073/pnas.0611503104
A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures
Abstract
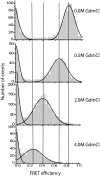
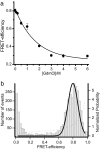
The yeast prion protein Sup35 is a translation termination factor, whose activity is modulated by sequestration into a self-perpetuating amyloid. The prion-determining domain, NM, consists of two distinct regions: an amyloidogenic N terminus domain (N) and a charged solubilizing middle region (M). To gain insight into prion conversion, we used single-molecule fluorescence resonance energy transfer (SM-FRET) and fluorescence correlation spectroscopy to investigate the structure and dynamics of monomeric NM. Low protein concentrations in these experiments prevented the formation of obligate on-pathway oligomers, allowing us to study early folding intermediates in isolation from higher-order species. SM-FRET experiments on a dual-labeled amyloid core variant (N21C/S121C, retaining wild-type prion behavior) indicated that the N region of NM adopts a collapsed form similar to "burst-phase" intermediates formed during the folding of many globular proteins, even though it lacks a typical hydrophobic core. The mean distance between residues 21 and 121 was approximately equal to 43 A. This increased with denaturant in a noncooperative fashion to approximately equal to 63 A, suggesting a multitude of interconverting species rather than a small number of discrete monomeric conformers. Fluorescence correlation spectroscopy analysis of singly labeled NM revealed fast conformational fluctuations on the 20- to 300-ns time scale. Quenching from proximal and distal tyrosines resulted in distinct fast and slower fluctuations. Our results indicate that native monomeric NM is composed of an ensemble of structures, having a collapsed and rapidly fluctuating N region juxtaposed with a more extended M region. The stability of such ensembles is likely to play a key role in prion conversion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

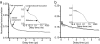
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

