Comparative proteomic and transcriptomic profiling of the fission yeast Schizosaccharomyces pombe
- PMID: 17299416
- PMCID: PMC1828747
- DOI: 10.1038/msb4100117
Comparative proteomic and transcriptomic profiling of the fission yeast Schizosaccharomyces pombe
Abstract
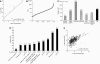
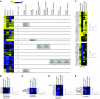
The fission yeast Schizosaccharomyces pombe is a widely used model organism to study basic mechanisms of eukaryotic biology, but unlike other model organisms, its proteome remains largely uncharacterized. Using a shotgun proteomics approach based on multidimensional prefractionation and tandem mass spectrometry, we have detected approximately 30% of the theoretical fission yeast proteome. Applying statistical modelling to normalize spectral counts to the number of predicted tryptic peptides, we have performed label-free quantification of 1465 proteins. The fission yeast protein data showed considerable correlations with mRNA levels and with the abundance of orthologous proteins in budding yeast. Functional pathway analysis indicated that the mRNA-protein correlation is strong for proteins involved in signalling and metabolic processes, but increasingly discordant for components of protein complexes, which clustered in groups with similar mRNA-protein ratios. Self-organizing map clustering of large-scale protein and mRNA data from fission and budding yeast revealed coordinate but not always concordant expression of components of functional pathways and protein complexes. This finding reaffirms at the protein level the considerable divergence in gene expression patterns of the two model organisms that was noticed in previous transcriptomic studies.
Figures



References
-
- Anderson NL, Matheson AD, Steiner S (2000) Proteomics: applications in basic and applied biology. Curr Opin Biotechnol 11: 408–412 - PubMed
-
- Bakhtiar R, Tse FL (2000) Biological mass spectrometry: a primer. Mutagenesis 15: 415–430 - PubMed
-
- Berriz GF, King OD, Bryant B, Sander C, Roth FP (2003) Characterizing gene sets with FuncAssociate. Bioinformatics 19: 2502–2504 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

