Mice develop normally in the absence of Smad4 nucleocytoplasmic shuttling
- PMID: 17300215
- PMCID: PMC1868808
- DOI: 10.1042/BJ20061830
Mice develop normally in the absence of Smad4 nucleocytoplasmic shuttling
Abstract
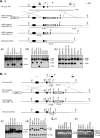
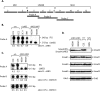
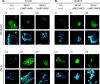
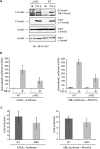
Smad4 in partnership with R-Smads (receptor-regulated Smads) activates TGF-beta (transforming growth factor-beta)-dependent signalling pathways essential for early mouse development. Smad4 null embryos die shortly after implantation due to severe defects in cell proliferation and visceral endoderm differentiation. In the basal state, Smad4 undergoes continuous shuttling between the cytoplasm and the nucleus due to the combined activities of an N-terminal NLS (nuclear localization signal) and an NES (nuclear export signal) located in its linker region. Cell culture experiments suggest that Smad4 nucleocytoplasmic shuttling plays an important role in TGF-beta signalling. In the present study we have investigated the role of Smad4 shuttling in vivo using gene targeting to engineer two independent mutations designed to eliminate Smad4 nuclear export. As predicted this results in increased levels of Smad4 in the nucleus of homozygous ES cells (embryonic stem cells) and primary keratinocytes, in the presence or absence of ligand. Neither mutation affects Smad4 expression levels nor its ability to mediate transcriptional activation in homozygous cell lines. Remarkably mouse mutants lacking the Smad4 NES develop normally. Smad4 NES mutants carrying one copy of a Smad4 null allele also fail to display developmental defects. The present study clearly demonstrates that Smad4 nucleocytoplasmic shuttling is not required for embryonic development or tissue homoeostasis in normal, healthy adult mice.
Figures

Similar articles
-
Transforming growth factor beta-independent shuttling of Smad4 between the cytoplasm and nucleus.Mol Cell Biol. 2000 Dec;20(23):9041-54. doi: 10.1128/MCB.20.23.9041-9054.2000. Mol Cell Biol. 2000. PMID: 11074002 Free PMC article.
-
An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity.Oncogene. 2003 Feb 20;22(7):1057-69. doi: 10.1038/sj.onc.1206212. Oncogene. 2003. PMID: 12592392
-
Analysis of Smad nucleocytoplasmic shuttling in living cells.J Cell Sci. 2004 Aug 15;117(Pt 18):4113-25. doi: 10.1242/jcs.01289. Epub 2004 Jul 27. J Cell Sci. 2004. PMID: 15280432
-
Nucleocytoplasmic shuttling of Smad proteins.Cell Res. 2009 Jan;19(1):36-46. doi: 10.1038/cr.2008.325. Cell Res. 2009. PMID: 19114992 Review.
-
The role of TGF-β/SMAD4 signaling in cancer.Int J Biol Sci. 2018 Jan 12;14(2):111-123. doi: 10.7150/ijbs.23230. eCollection 2018. Int J Biol Sci. 2018. PMID: 29483830 Free PMC article. Review.
Cited by
-
Dynamics of TGF-β signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor Smad4.Proc Natl Acad Sci U S A. 2012 Jul 10;109(28):E1947-56. doi: 10.1073/pnas.1207607109. Epub 2012 Jun 11. Proc Natl Acad Sci U S A. 2012. PMID: 22689943 Free PMC article.
-
Effect of interrupted endogenous BMP/Smad signaling on growth and steroidogenesis of porcine granulosa cells.J Zhejiang Univ Sci B. 2010 Sep;11(9):719-27. doi: 10.1631/jzus.B1000079. J Zhejiang Univ Sci B. 2010. PMID: 20803776 Free PMC article.
-
Morphogen interpretation: concentration, time, competence, and signaling dynamics.Wiley Interdiscip Rev Dev Biol. 2017 Jul;6(4):e271. doi: 10.1002/wdev.271. Epub 2017 Mar 20. Wiley Interdiscip Rev Dev Biol. 2017. PMID: 28319331 Free PMC article. Review.
-
IκBα Nuclear Export Enables 4-1BB-Induced cRel Activation and IL-2 Production to Promote CD8 T Cell Immunity.J Immunol. 2020 Sep 15;205(6):1540-1553. doi: 10.4049/jimmunol.2000039. Epub 2020 Aug 14. J Immunol. 2020. PMID: 32817348 Free PMC article.
-
Wip1 regulates Smad4 phosphorylation and inhibits TGF-β signaling.EMBO Rep. 2020 May 6;21(5):e48693. doi: 10.15252/embr.201948693. Epub 2020 Feb 27. EMBO Rep. 2020. PMID: 32103600 Free PMC article.
References
-
- Shi Y., Massague J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. - PubMed
-
- Massague J., Seoane J., Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. - PubMed
-
- Reguly T., Wrana J. L. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends Cell Biol. 2003;13:216–220. - PubMed
-
- Xu L., Kang Y., Col S., Massague J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell. 2002;10:271–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

