Structural definition of a conserved neutralization epitope on HIV-1 gp120
- PMID: 17301785
- PMCID: PMC2584968
- DOI: 10.1038/nature05580
Structural definition of a conserved neutralization epitope on HIV-1 gp120
Abstract
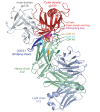
The remarkable diversity, glycosylation and conformational flexibility of the human immunodeficiency virus type 1 (HIV-1) envelope (Env), including substantial rearrangement of the gp120 glycoprotein upon binding the CD4 receptor, allow it to evade antibody-mediated neutralization. Despite this complexity, the HIV-1 Env must retain conserved determinants that mediate CD4 binding. To evaluate how these determinants might provide opportunities for antibody recognition, we created variants of gp120 stabilized in the CD4-bound state, assessed binding of CD4 and of receptor-binding-site antibodies, and determined the structure at 2.3 A resolution of the broadly neutralizing antibody b12 in complex with gp120. b12 binds to a conformationally invariant surface that overlaps a distinct subset of the CD4-binding site. This surface is involved in the metastable attachment of CD4, before the gp120 rearrangement required for stable engagement. A site of vulnerability, related to a functional requirement for efficient association with CD4, can therefore be targeted by antibody to neutralize HIV-1.
Conflict of interest statement
Coordinates and structure factors have been deposited in the Protein Data Bank and may be obtained from the authors (accession codes 2nxy–2ny6 for the nine variant gp120 molecules with CD4 and 17b; accession code 2ny7 for the b12–gp120 complex). Reprints and permissions information is available at
Figures



Comment in
-
HIV-1 rational vaccine design: molecular details of b12-gp120 complex structure.Expert Rev Vaccines. 2007 Jun;6(3):319-21. doi: 10.1586/14760584.6.3.319. Expert Rev Vaccines. 2007. PMID: 17542747 No abstract available.
References
-
- Joint United National Programme on HIV/AIDS. 2006 Report on the global AIDS epidemic. 〈http://www.aids.org/en/HIVdata/2006GlobalReport/〉 (2006)
-
- Parren PW, Moore JP, Burton DR, Sattentua QJ. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13:S137–S162. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

