Ancient origin of the new developmental superfamily DANGER
- PMID: 17301879
- PMCID: PMC1784063
- DOI: 10.1371/journal.pone.0000204
Ancient origin of the new developmental superfamily DANGER
Abstract
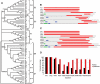
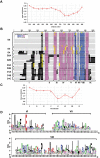
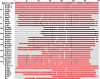
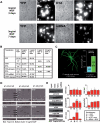
Developmental proteins play a pivotal role in the origin of animal complexity and diversity. We report here the identification of a highly divergent developmental protein superfamily (DANGER), which originated before the emergence of animals (approximately 850 million years ago) and experienced major expansion-contraction events during metazoan evolution. Sequence analysis demonstrates that DANGER proteins diverged via multiple mechanisms, including amino acid substitution, intron gain and/or loss, and recombination. Divergence for DANGER proteins is substantially greater than for the prototypic member of the superfamily (Mab-21 family) and other developmental protein families (e.g., WNT proteins). DANGER proteins are widely expressed and display species-dependent tissue expression patterns, with many members having roles in development. DANGER1A, which regulates the inositol trisphosphate receptor, promotes the differentiation and outgrowth of neuronal processes. Regulation of development may be a universal function of DANGER family members. This family provides a model system to investigate how rapid protein divergence contributes to morphological complexity.
Conflict of interest statement
Figures


References
-
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Malden, MA: Blackwell Publishers; 2004.
-
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. - PubMed
-
- Avise JC. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard University Press; 2000.
-
- van Rossum DB, Patterson RL, Cheung KH, Barrow RK, Syrovatkina V, et al. DANGER, a novel regulatory protein of inositol 1,4,5-trisphosphate-receptor activity. J Biol Chem. 2006;281:37111–37116. - PubMed
-
- Chow KL, Hall DH, Emmons SW. The mab-21 gene of Caenorhabditis elegans encodes a novel protein required for choice of alternate cell fates. Development. 1995;121:3615–3626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

