Regulation of oncogenic transcription factor hTAF(II)68-TEC activity by human glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
- PMID: 17302560
- PMCID: PMC1868794
- DOI: 10.1042/BJ20061297
Regulation of oncogenic transcription factor hTAF(II)68-TEC activity by human glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
Abstract
Tumour-specific chromosomal rearrangements are known to create chimaeric products with the ability to generate many human cancers. hTAF(II)68-TEC (where hTAF(II)68 is human TATA-binding protein-associated factor II 68 and TEC is translocated in extraskeletal chondrosarcoma) is such a fusion product, resulting from a t(9;17) chromosomal translocation found in extraskeletal myxoid chondrosarcomas, where the hTAF(II)68 NTD (N-terminal domain) is fused to TEC protein. To identify proteins that control hTAF(II)68-TEC function, we used affinity chromatography on immobilized hTAF(II)68 (NTD) and MALDI-TOF (matrix-assisted laser-desorption ionization-time-of-flight) MS and isolated a novel hTAF(II)68-TEC-interacting protein, GAPDH (glyceraldehyde-3-phosphate dehydrogenase). GAPDH is a glycolytic enzyme that is also involved in the early steps of apoptosis, nuclear tRNA export, DNA replication, DNA repair and transcription. hTAF(II)68-TEC and GAPDH were co-immunoprecipitated from cell extracts, and glutathione S-transferase pull-down assays revealed that the C-terminus of hTAF(II)68 (NTD) was required for interaction with GAPDH. In addition, three independent regions of GAPDH (amino acids 1-66, 67-160 and 160-248) were involved in binding to hTAF(II)68 (NTD). hTAF(II)68-TEC-dependent transcription was enhanced by GAPDH, but not by a GAPDH mutant defective in hTAF(II)68-TEC binding. Moreover, a fusion of GAPDH with the GAL4 DNA-binding domain increased the promoter activity of a reporter containing GAL4 DNA-binding sites, demonstrating the presence of a transactivation domain(s) in GAPDH. The results of the present study suggest that the transactivation potential of the hTAF(II)68-TEC oncogene product is positively modulated by GAPDH.
Figures


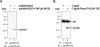





References
-
- Clark J., Benjamin H., Gill S., Sidhar S., Goodwin G., Crew J., Gusterson B. A., Shipley J., Cooper C. S. Fusion of the EWS gene to CHN, a member of the steroid/thyroid receptor gene superfamily, in a human myxoid chondrosarcoma. Oncogene. 1996;12:229–235. - PubMed
-
- Labelle Y., Zucman J., Stenman G., Kindblom L. G., Knight J., Turc-Carel C., Dockhorn-Dworniczak B., Mandahl N., Desmaze C., Peter M., et al. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum. Mol. Genet. 1995;4:2219–2226. - PubMed
-
- Labelle Y., Bussieres J., Courjal F., Goldring M. B. The EWS/TEC fusion protein encoded by the t(9;22) chromosomal translocation in human chondrosarcomas is a highly potent transcriptional activator. Oncogene. 1999;18:3303–3308. - PubMed
-
- Sjogren H., Meis-Kindblom J., Kindblom L. G., Aman P., Stenman G. Fusion of the EWS-related gene TAF2N to TEC in extraskeletal myxoid chondrosarcoma. Cancer Res. 1999;59:5064–5067. - PubMed
-
- Attwooll C., Tariq M., Harris M., Coyne J. D., Telford N., Varley J. M. Identification of a novel fusion gene involving hTAFII68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene. 1999;18:7599–7601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

