Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes
- PMID: 17304215
- PMCID: PMC1817630
- DOI: 10.1038/sj.emboj.7601582
Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes
Abstract
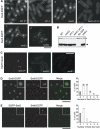
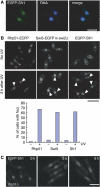
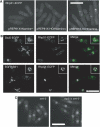
Several accessory proteins referred to as mediators are required for the full activity of the Rad51 (Rhp51 in fission yeast) recombinase. In this study, we analyzed in vivo functions of the recently discovered Swi5/Sfr1 complex from fission yeast. In normally growing cells, the Swi5-GFP protein localizes to the nucleus, where it forms a diffuse nuclear staining pattern with a few distinct foci. These spontaneous foci do not form in swi2Delta mutants. Upon UV irradiation, Swi5 focus formation is induced in swi2Delta mutants, a response that depends on Sfr1 function, and Sfr1 also forms foci that colocalize with damage-induced Rhp51 foci. The number of UV-induced Rhp51 foci is partially reduced in swi5Delta and rhp57Delta mutants and completely abolished in an swi5Delta rhp57Delta double mutant. An assay for products generated by HO endonuclease-induced DNA double-strand breaks (DSBs) reveals that Rhp51 and Rhp57, but not Swi5/Sfr1, are essential for crossover production. These results suggest that Swi5/Sfr1 functions as an Rhp51 mediator but processes DSBs in a manner different from that of the Rhp55/57 mediator.
Figures


References
-
- Allers T, Lichten M (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 - PubMed
-
- Arcangioli B, Thon G (2004) Mating-type cassettes: structure, switching and silencing. In The Molecular Biology of Schizosaccharomyces pombe, Egal R (ed) pp 129–148. Heidelberg: Springer
-
- Bishop DK, Park D, Xu L, Kleckner N (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

