Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets
- PMID: 17304354
- PMCID: PMC1794115
- DOI: 10.1172/JCI26833
Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets
Abstract
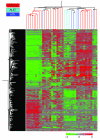
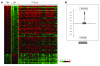
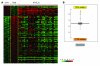
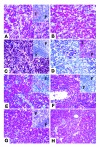
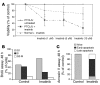
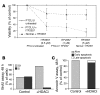
Peripheral T cell lymphoma, unspecified (PTCL/U), the most common form of PTCL, displays heterogeneous morphology and phenotype, poor response to treatment, and poor prognosis. We demonstrate that PTCL/U shows a gene expression profile clearly distinct from that of normal T cells. Comparison with the profiles of purified T cell subpopulations (CD4+, CD8+, resting [HLA-DR-], and activated [HLA-DR+]) reveals that PTCLs/U are most closely related to activated peripheral T lymphocytes, either CD4+ or CD8+. Interestingly, the global gene expression profile cannot be surrogated by routine CD4/CD8 immunohistochemistry. When compared with normal T cells, PTCLs/U display deregulation of functional programs often involved in tumorigenesis (e.g., apoptosis, proliferation, cell adhesion, and matrix remodeling). Products of deregulated genes can be detected in PTCLs/U by immunohistochemistry with an ectopic, paraphysiologic, or stromal location. PTCLs/U aberrantly express, among others, PDGFRalpha, a tyrosine-kinase receptor, whose deregulation is often related to a malignant phenotype. Notably, both phosphorylation of PDGFRalpha and sensitivity of cultured PTCL cells to imatinib (as well as to an inhibitor of histone deacetylase) were found. These results, which might be extended to other more rare PTCL categories, provide insight into tumor pathogenesis and clinical management of PTCL/U.
Figures


References
-
- Jaffe, E.S., and Ralfkiaer, E. 2001. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. IARC Press. Lyon, France. 191–194.
-
- Kadin M.E., Berard C.W., Nanba K., Wakasa H. Lymphoproliferative diseases in Japan and Western countries: Proceedings of the United States–Japan Seminar, September 6 and 7, 1982, in Seattle, Washington. Hum. Pathol. 1983;14:745–772. - PubMed
-
- Harris N.L., et al. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 1994;84:1361–1392. - PubMed
-
- Lopez-Guillermo A., et al. Peripheral T-cell lymphomas: initial features, natural history, and prognostic factors in a series of 174 patients diagnosed according to the R.E.A.L. Classification. Ann. Oncol. 1998;9:849–855. - PubMed
-
- Rizvi M.A., Evens A.M., Tallman M.S., Nelson B.P., Rosen S.T. T-cell non-Hodgkin lymphoma. Blood. 2006;107:1255–1264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

