Functional impact of missense variants in BRCA1 predicted by supervised learning
- PMID: 17305420
- PMCID: PMC1797820
- DOI: 10.1371/journal.pcbi.0030026
Functional impact of missense variants in BRCA1 predicted by supervised learning
Abstract
Many individuals tested for inherited cancer susceptibility at the BRCA1 gene locus are discovered to have variants of unknown clinical significance (UCVs). Most UCVs cause a single amino acid residue (missense) change in the BRCA1 protein. They can be biochemically assayed, but such evaluations are time-consuming and labor-intensive. Computational methods that classify and suggest explanations for UCV impact on protein function can complement functional tests. Here we describe a supervised learning approach to classification of BRCA1 UCVs. Using a novel combination of 16 predictive features, the algorithms were applied to retrospectively classify the impact of 36 BRCA1 C-terminal (BRCT) domain UCVs biochemically assayed to measure transactivation function and to blindly classify 54 documented UCVs. Majority vote of three supervised learning algorithms is in agreement with the assay for more than 94% of the UCVs. Two UCVs found deleterious by both the assay and the classifiers reveal a previously uncharacterized putative binding site. Clinicians may soon be able to use computational classifiers such as those described here to better inform patients. These classifiers can be adapted to other cancer susceptibility genes and systematically applied to prioritize the growing number of potential causative loci and variants found by large-scale disease association studies.
Conflict of interest statement
Figures

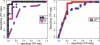

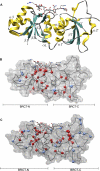
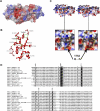
References
-
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. - PubMed
-
- Starita LM, Parvin JD. Substrates of the BRCA1-dependent ubiquitin ligase. Cancer Biol Ther. 2006;5:137–141. - PubMed
-
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. - PubMed
-
- Nathanson KN, Wooster R, Weber BL. Breast cancer genetics: What we know and what we need. Nat Med. 2001;7:552–556. - PubMed
-
- Szabo CI, Worley T, Monteiro AN. Understanding germ-line mutations in BRCA1. Cancer Biol Ther. 2004;3:515–520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

