An evaluation of LSU rDNA D1-D2 sequences for their use in species identification
- PMID: 17306026
- PMCID: PMC1805435
- DOI: 10.1186/1742-9994-4-6
An evaluation of LSU rDNA D1-D2 sequences for their use in species identification
Abstract
Background: Identification of species via DNA sequences is the basis for DNA taxonomy and DNA barcoding. Currently there is a strong focus on using a mitochondrial marker for this purpose, in particular a fragment from the cytochrome oxidase I gene (COI). While there is ample evidence that this marker is indeed suitable across a broad taxonomic range to delineate species, it has also become clear that a complementation by a nuclear marker system could be advantageous. Ribosomal RNA genes could be suitable for this purpose, because of their global occurrence and the possibility to design universal primers. However, it has so far been assumed that these genes are too highly conserved to allow resolution at, or even beyond the species level. On the other hand, it is known that ribosomal gene regions harbour also highly divergent parts. We explore here the information content of two adjacent divergence regions of the large subunit ribosomal gene, the D1-D2 region.
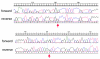
Results: Universal primers were designed to amplify the D1-D2 region from all metazoa. We show that amplification products in the size between 800-1300 bp can be obtained across a broad range of animal taxa, provided some optimizations of the PCR procedure are implemented. Although the ribosomal genes occur in multiple copies in the genomes, we find generally very little intra-individual polymorphism (<< 0.1% on average) indicating that concerted evolution is very effective in most cases. Studies in two fish taxa (genus Cottus and genus Aphyosemion) show that the D1-D2 LSU sequence can resolve even very closely related species with the same fidelity as COI sequences. In one case we can even show that a mitochondrial transfer must have occurred, since the nuclear sequence confirms the taxonomic assignment, while the mitochondrial sequence would have led to the wrong classification. We have further explored whether hybrids between species can be detected with the nuclear sequence and we show for a test case of natural hybrids among cyprinid fish species (Alburnus alburnus and Rutilus rutilus) that this is indeed possible.
Conclusion: The D1-D2 LSU region is a suitable marker region for applications in DNA based species identification and should be considered to be routinely used as a marker complementing broad scale studies based on mitochondrial markers.
Figures





Similar articles
-
The D1-D2 region of the large subunit ribosomal DNA as barcode for ciliates.Mol Ecol Resour. 2014 May;14(3):458-68. doi: 10.1111/1755-0998.12195. Epub 2013 Nov 29. Mol Ecol Resour. 2014. PMID: 24165195
-
A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents.Front Zool. 2013 Jun 14;10:34. doi: 10.1186/1742-9994-10-34. eCollection 2013. Front Zool. 2013. PMID: 23767809 Free PMC article.
-
Evaluation of ribosomal RNA and actin gene sequences for the identification of ascomycetous yeasts.Int J Food Microbiol. 2003 Sep 1;86(1-2):61-78. doi: 10.1016/s0168-1605(03)00248-4. Int J Food Microbiol. 2003. PMID: 12892922
-
Takashi Nakase's last tweet: what is the current direction of microbial taxonomy research?FEMS Yeast Res. 2019 Dec 1;19(8):foz066. doi: 10.1093/femsyr/foz066. FEMS Yeast Res. 2019. PMID: 31816016 Review.
-
Ribosomal DNA: molecular evolution and phylogenetic inference.Q Rev Biol. 1991 Dec;66(4):411-53. doi: 10.1086/417338. Q Rev Biol. 1991. PMID: 1784710 Review.
Cited by
-
SNP typing for germplasm identification of Amomum villosum Lour. Based on DNA barcoding markers.PLoS One. 2014 Dec 22;9(12):e114940. doi: 10.1371/journal.pone.0114940. eCollection 2014. PLoS One. 2014. PMID: 25531885 Free PMC article.
-
Integrative species delimitation in photosynthetic sea slugs reveals twenty candidate species in three nominal taxa studied for drug discovery, plastid symbiosis or biological control.Mol Phylogenet Evol. 2013 Dec;69(3):1101-19. doi: 10.1016/j.ympev.2013.07.009. Epub 2013 Jul 19. Mol Phylogenet Evol. 2013. PMID: 23876292 Free PMC article.
-
A microbial eukaryote with a unique combination of purple bacteria and green algae as endosymbionts.Sci Adv. 2021 Jun 11;7(24):eabg4102. doi: 10.1126/sciadv.abg4102. Print 2021 Jun. Sci Adv. 2021. PMID: 34117067 Free PMC article.
-
Hidden biodiversity revealed by integrated morphology and genetic species delimitation of spring dwelling water mite species (Acari, Parasitengona: Hydrachnidia).Parasit Vectors. 2019 Oct 21;12(1):492. doi: 10.1186/s13071-019-3750-y. Parasit Vectors. 2019. PMID: 31639027 Free PMC article.
-
A genome-skimmed phylogeny of a widespread bryozoan family, Adeonidae.BMC Evol Biol. 2019 Dec 27;19(1):235. doi: 10.1186/s12862-019-1563-4. BMC Evol Biol. 2019. PMID: 31881939 Free PMC article.
References
-
- Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AP. A plea for DNA taxonomy. Trends in Ecology & Evolution. 2003;18:70–74. doi: 10.1016/S0169-5347(02)00041-1. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

