Relationship of pulsed-field profiles with key phage types of Salmonella enterica serotype Enteritidis in Europe: results of an international multi-centre study
- PMID: 17306053
- PMCID: PMC2870705
- DOI: 10.1017/S0950268807008102
Relationship of pulsed-field profiles with key phage types of Salmonella enterica serotype Enteritidis in Europe: results of an international multi-centre study
Abstract
Salmonella is one of the most common causes of foodborne infection in Europe with Salmonella enterica serovar Enteritidis (S. Enteritidis) being the most commonly identified serovar. The predominant phage type for S. Enteritidis is phage type (PT) 4, although PT 8 has increased in incidence. Within these phage types, pulsed-field gel electrophoresis (PFGE) provides a method of further subdivision. The international project, Salm-gene, was established in 2001 to develop a database of PFGE profiles within nine European countries and to establish criteria for real-time pattern recognition. It uses DNA fingerprints of salmonellas to investigate outbreaks and to evaluate trends and emerging issues of foodborne infection within Europe. The Salm-gene database contains details of about 11 700 S. Enteritidis isolates, demonstrating more than 65 unique PFGE profiles. The clonal nature of S. Enteritidis is evidenced by the high similarity and distribution of PFGE profiles. Over 56% (6603/11 716) of the submitted isolates of several different phage types were profile SENTXB.0001, although this profile is most closely associated with PT 4. The next most common profiles, SENTXB.0002 and SENTXB.0005, were closely associated with PT 8 and PT 21 respectively. Studies to investigate the relationship of profile types with outbreaks and possible vehicles of infection suggest that the incidence of PFGE profile SENTXB.0002, and thus PT 8, in some countries may be due to importation of foods or food production animals from Eastern Europe, where PT 8 is amongst the most frequently identified phage types. Collation of subtyping data, especially in the commonly recognized phage types, is necessary in order to evaluate trends and emerging issues in salmonella infection.
Figures

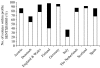
 , PT 6;
, PT 6; , PT 14b;
, PT 14b; , PT 21;
, PT 21; , PT 1.
, PT 1.
Similar articles
-
Distribution of molecular subtypes within Salmonella enterica serotype Enteritidis phage type 4 and S. Typhimurium definitive phage type 104 in nine European countries, 2000-2004: results of an international multi-centre study.Epidemiol Infect. 2006 Aug;134(4):729-36. doi: 10.1017/S0950268805005820. Epub 2006 Jan 25. Epidemiol Infect. 2006. PMID: 16436221 Free PMC article.
-
Pulsed-field gel electrophoresis, plasmid profiles and phage types for the human isolates of Salmonella enterica serovar Enteritidis obtained over 13 years in Taiwan.J Appl Microbiol. 2005;99(6):1472-83. doi: 10.1111/j.1365-2672.2005.02749.x. J Appl Microbiol. 2005. PMID: 16313420
-
Analysis of molecular epidemiology of Chilean Salmonella enterica serotype enteritidis isolates by pulsed-field gel electrophoresis and bacteriophage typing.J Clin Microbiol. 2003 Apr;41(4):1617-22. doi: 10.1128/JCM.41.4.1617-1622.2003. J Clin Microbiol. 2003. PMID: 12682153 Free PMC article.
-
Molecular typing methods for S. enteritidis.Int J Food Microbiol. 1994 Jan;21(1-2):69-77. doi: 10.1016/0168-1605(94)90201-1. Int J Food Microbiol. 1994. PMID: 7908822 Review.
-
Molecular epidemiological techniques for Salmonella strain discrimination.Front Biosci. 2003 Jan 1;8:c14-24. doi: 10.2741/947. Front Biosci. 2003. PMID: 12456335 Review.
Cited by
-
Genomic and phenotypic variation in epidemic-spanning Salmonella enterica serovar Enteritidis isolates.BMC Microbiol. 2009 Nov 18;9:237. doi: 10.1186/1471-2180-9-237. BMC Microbiol. 2009. PMID: 19922635 Free PMC article.
-
Comparative study of all Salmonella enterica serovar Enteritidis strains isolated from food and food animals in Greece from 2008 to 2010 with clinical isolates.Eur J Clin Microbiol Infect Dis. 2016 May;35(5):741-6. doi: 10.1007/s10096-016-2591-2. Epub 2016 Feb 10. Eur J Clin Microbiol Infect Dis. 2016. PMID: 26864044
-
Complete Genome and Plasmid Sequences of Three Canadian Strains of Salmonella enterica subsp. enterica Serovar Enteritidis Belonging to Phage Types 8, 13, and 13a.Genome Announc. 2015 Sep 24;3(5):e01017-15. doi: 10.1128/genomeA.01017-15. Genome Announc. 2015. PMID: 26404595 Free PMC article.
-
A multi-country outbreak of Salmonella Newport gastroenteritis in Europe associated with watermelon from Brazil, confirmed by whole genome sequencing: October 2011 to January 2012.Euro Surveill. 2014 Aug 7;19(31):6-13. doi: 10.2807/1560-7917.es2014.19.31.20866. Euro Surveill. 2014. PMID: 25138971 Free PMC article.
-
Evaluation of the use of CRISPR loci for discrimination of Salmonella enterica subsp. enterica serovar Enteritidis strains recovered in Canada and comparison with other subtyping methods.AIMS Microbiol. 2022 Jul 15;8(3):300-317. doi: 10.3934/microbiol.2022022. eCollection 2022. AIMS Microbiol. 2022. PMID: 36317002 Free PMC article.
References
-
- Anon 2005. http://www.efsa.eu.int/science/monitoring_zoonoses/reports/1277_en.html. http://www.efsa.eu.int/science/monitoring_zoonoses/reports/1277_en.html . European Food Safety Authority (EFSA): Zoonoses reports, ). Accessed 5 September 2006.
-
- Anon. EFSA Working group on opinion of the Scientific Panel on Biological Hazards related to the use of antimicrobials for the control of Salmonella in poultry. European Food Safety Authority Journal. 2004;115:1–76.
-
- Ward LR et al. A collaborative investigation of an outbreak of Salmonella enterica serotype Newport in England and Wales in 2001 associated with ready-to-eat salad vegetables. Communicable Disease & Public Health. 2002;5:301–304. - PubMed
-
- Fisher IST. The Enter-net international surveillance network – how it works. Eurosurveillance. 1999;4:52–55. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

