Global analysis of alternative splicing during T-cell activation
- PMID: 17307815
- PMCID: PMC1831861
- DOI: 10.1261/rna.457207
Global analysis of alternative splicing during T-cell activation
Abstract
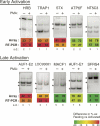
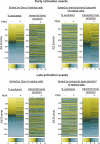
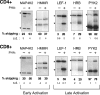
The role of alternative splicing (AS) in eliciting immune responses is poorly understood. We used quantitative AS microarray profiling to survey changes in AS during activation of Jurkat cells, a leukemia-derived T-cell line. Our results indicate that approximately 10-15% of the profiled alternative exons undergo a >10% change in inclusion level during activation. The majority of the genes displaying differential AS levels are distinct from the set of genes displaying differential transcript levels. These two gene sets also have overlapping yet distinct functional roles. For example, genes that show differential AS patterns during T-cell activation are often closely associated with cell-cycle regulation, whereas genes with differential transcript levels are highly enriched in functions associated more directly with immune defense and cytoskeletal architecture. Previously unknown AS events were detected in genes that have important roles in T-cell activation, and these AS level changes were also observed during the activation of normal human peripheral CD4+ and CD8+ lymphocytes. In summary, by using AS microarray profiling, we have discovered many new AS changes associated with T-cell activation. Our results suggest an extensive role for AS in the regulation of the mammalian immune response.
Figures

References
-
- Abraham, R.T., Weiss, A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol. 2004;4:301–308. - PubMed
-
- Beissbarth, T., Speed, T.P. GOstat: Find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. - PubMed
-
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. - PubMed
-
- Georgopoulos, K., Winandy, S., Avitahl, N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu. Rev. Immunol. 1997;15:155–176. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
