Measuring the folding transition time of single RNA molecules
- PMID: 17307831
- PMCID: PMC1852359
- DOI: 10.1529/biophysj.106.094623
Measuring the folding transition time of single RNA molecules
Abstract
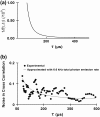
We describe a new, time-apertured photon correlation method for resolving the transition time between two states of RNA in folding--i.e., the time of the transition between states rather than the time spent in each state. Single molecule fluorescence resonance energy transfer and fluorescence correlation spectroscopy are used to obtain these measurements. Individual RNA molecules are labeled with fluorophores such as Cy3 and Cy5. Those molecules are then immobilized on a surface and observed for many seconds during which time the molecules spontaneously switch between two conformational states with different levels of flourescence resonance energy transfer efficiency. Single photons are counted from each fluorophore and cross correlated in a small window around a transition. The average of over 1000 cross correlations can be fit to a polynomial, which can determine transition times as short as the average photon emission interval. We applied the method to the P4-P6 domain of the Tetrahymena group I self-splicing intron to yield the folding transition time of 240 micros. The unfolding time is found to be too short to measure with this method.
Figures



Similar articles
-
Direct measurement of tertiary contact cooperativity in RNA folding.J Am Chem Soc. 2008 May 14;130(19):6085-7. doi: 10.1021/ja800919q. Epub 2008 Apr 23. J Am Chem Soc. 2008. PMID: 18429611 Free PMC article.
-
The structure and folding of branched RNA analyzed by fluorescence resonance energy transfer.Methods Enzymol. 2009;469:159-87. doi: 10.1016/S0076-6879(09)69008-X. Methods Enzymol. 2009. PMID: 20946789
-
An early transition state for folding of the P4-P6 RNA domain.RNA. 2001 Feb;7(2):161-6. doi: 10.1017/s1355838201001716. RNA. 2001. PMID: 11233973 Free PMC article.
-
Probing the kinetic and thermodynamic consequences of the tetraloop/tetraloop receptor monovalent ion-binding site in P4-P6 RNA by smFRET.Biochem Soc Trans. 2015 Apr;43(2):172-8. doi: 10.1042/BST20140268. Biochem Soc Trans. 2015. PMID: 25849913 Free PMC article. Review.
-
Transition Path Times Measured by Single-Molecule Spectroscopy.J Mol Biol. 2018 Feb 16;430(4):409-423. doi: 10.1016/j.jmb.2017.05.018. Epub 2017 May 25. J Mol Biol. 2018. PMID: 28551335 Free PMC article. Review.
Cited by
-
Recent Developments in Nanosensors for Imaging Applications in Biological Systems.Annu Rev Anal Chem (Palo Alto Calif). 2019 Jun 12;12(1):109-128. doi: 10.1146/annurev-anchem-061417-125747. Epub 2019 Mar 11. Annu Rev Anal Chem (Palo Alto Calif). 2019. PMID: 30857408 Free PMC article. Review.
-
Microsecond folding experiments and simulations: a match is made.Phys Chem Chem Phys. 2013 Mar 14;15(10):3372-88. doi: 10.1039/c3cp43992e. Epub 2013 Jan 29. Phys Chem Chem Phys. 2013. PMID: 23361200 Free PMC article.
-
RNA folding: conformational statistics, folding kinetics, and ion electrostatics.Annu Rev Biophys. 2008;37:197-214. doi: 10.1146/annurev.biophys.37.032807.125957. Annu Rev Biophys. 2008. PMID: 18573079 Free PMC article. Review.
-
Structure-function relationship of substituted bromomethylcoumarins in nucleoside specificity of RNA alkylation.PLoS One. 2013 Jul 3;8(7):e67945. doi: 10.1371/journal.pone.0067945. Print 2013. PLoS One. 2013. PMID: 23844135 Free PMC article.
-
Kinetic and thermodynamic framework for P4-P6 RNA reveals tertiary motif modularity and modulation of the folding preferred pathway.Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):E4956-65. doi: 10.1073/pnas.1525082113. Epub 2016 Aug 4. Proc Natl Acad Sci U S A. 2016. PMID: 27493222 Free PMC article.
References
-
- Ha, T., I. Rasnik, W. Cheng, H. P. Babcock, G. H. Gauss, T. M. Lohman, and S. Chu. 2002. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 419:638–641. - PubMed
-
- Weiss, S. 2000. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 7:724–729. - PubMed
-
- Babcock, H., X. W. Zhuang, R. Russell, L. Bartley, D. Herschlag, and S. Chu. 2001. A single molecule study of the folding dynamics of a large RNA enzyme. Biophys. J. 80:201A. (Abstr.).
-
- Zhuang, X., L. E. Bartley, H. P. Babcock, R. Russell, T. Ha, D. Herschlag, and S. Chu. 2000. A single-molecule study of RNA catalysis and folding. Science. 288:2048–2051. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

