Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate
- PMID: 17307853
- PMCID: PMC1855889
- DOI: 10.1128/JB.01940-06
Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate
Abstract
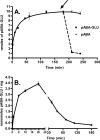
Escherichia coli AbgT was first identified as a structural protein enabling the growth of p-aminobenzoate auxotrophs on exogenous p-aminobenzoyl-glutamate (M. J. Hussein, J. M. Green, and B. P. Nichols, J. Bacteriol. 180:6260-6268, 1998). The abg region includes abgA, abgB, abgT, and ogt; these genes may be regulated by AbgR, a divergently transcribed LysR-type protein. Wild-type cells transformed with a high-copy-number plasmid encoding abgT demonstrate saturable uptake of p-aminobenzoyl-glutamate (K(T)=123 microM); control cells expressing vector demonstrate negligible uptake. The addition of metabolic poisons inhibited uptake of p-aminobenzoyl-glutamate, consistent with this process requiring energy. p-Aminobenzoyl-glutamate taken in by cells expressing large amounts of AbgT alone is not rapidly metabolized to a form that is trapped in the cell, as the addition of nonradioactive p-aminobenzoyl-glutamate to these cells results in a rapid loss of intracellular label. The addition of nonradioactive p-aminobenzoate has no effect. The abgA, abgB, and abgAB genes were cloned into the medium-copy-number plasmid pACYC184; p-aminobenzoate auxotrophs transformed with the clone encoding abgAB demonstrated enhanced ability to grow on low levels of p-aminobenzoyl-glutamate. When transformed with complementary plasmids encoding high-copy levels of abgT and medium-copy levels of abgAB, p-aminobenzoate auxotrophs grew on 50 nM p-aminobenzoyl-glutamate. Our data are consistent with a model of p-aminobenzoyl-glutamate utilization in which AbgT catalyzes transport of p-aminobenzoyl-glutamate, followed by cleavage to p-aminobenzoate by a protein composed of subunits encoded by abgA and abgB. While endogenous expression of these genes is very low under the conditions in which we performed our experiments, these genes may be induced by AbgR bound to an unknown molecule. The true physiological role of this region may be related to some molecule similar to p-aminobenzoyl-glutamate, such as a dipeptide.
Figures



References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Short protocols in molecular biology, 4th ed. John Wiley & Sons, Inc., New York, NY.
-
- Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
-
- Green, J. M., B. P. Nichols, and R. G. Matthews. 1996. Folate biosynthesis, reduction, and polyglutamylation, p. 665-673. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. I. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

