Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms
- PMID: 17307901
- PMCID: PMC1851817
- DOI: 10.1104/pp.106.092536
Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms
Abstract
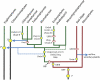
The light-harvesting complexes (LHCs) of land plants and green algae have essential roles in light capture and photoprotection. Though the functional diversity of the individual LHC proteins are well described in many land plants, the extent of this family in the majority of green algal groups is unknown. To examine the evolution of the chlorophyll a/b antennae system and to infer its ancestral state, we initiated several expressed sequence tag projects from a taxonomically broad range of chlorophyll a/b-containing protists. This included representatives from the Ulvophyceae (Acetabularia acetabulum), the Mesostigmatophyceae (Mesostigma viride), and the Prasinophyceae (Micromonas sp.), as well as one representative from each of the Euglenozoa (Euglena gracilis) and Chlorarachniophyta (Bigelowiella natans), whose plastids evolved secondarily from a green alga. It is clear that the core antenna system was well developed prior to green algal diversification and likely consisted of the CP29 (Lhcb4) and CP26 (Lhcb5) proteins associated with photosystem II plus a photosystem I antenna composed of proteins encoded by at least Lhca3 and two green algal-specific proteins encoded by the Lhca2 and 9 genes. In organisms containing secondary plastids, we found no evidence for orthologs to the plant/algal antennae with the exception of CP29. We also identified PsbS homologs in the Ulvophyceae and the Prasinophyceae, indicating that this distinctive protein appeared prior to green algal diversification. This analysis provides a snapshot of the antenna systems in diverse green algae, and allows us to infer the changing complexity of the antenna system during green algal evolution.
Figures



References
-
- Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21 2104–2105 - PubMed
-
- Akaike H (1974) New look at statistical-model identification. IEEE Transactions on Automatic Control 19 716–723
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

