Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds
- PMID: 17307924
- PMCID: PMC1867329
- DOI: 10.1105/tpc.106.048934
Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds
Abstract
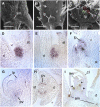
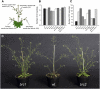
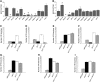
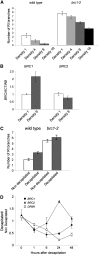
Shoot branching patterns depend on a key developmental decision: whether axillary buds grow out to give a branch or whether they remain dormant in the axils of leaves. This decision is controlled by endogenous and environmental stimuli mediated by hormonal signals. Although genes involved in the long-distance signaling of this process have been identified, the genes responding inside the buds to cause growth arrest remained unknown in Arabidopsis thaliana. Here, we describe an Arabidopsis gene encoding a TCP transcription factor closely related to teosinte branched1 (tb1) from maize (Zea mays), BRANCHED1 (BRC1), which represents a key point at which signals controlling branching are integrated within axillary buds. BRC1 is expressed in developing buds, where it arrests bud development. BRC1 downregulation leads to branch outgrowth. BRC1 responds to developmental and environmental stimuli controlling branching and mediates the response to these stimuli. Mutant and expression analyses suggest that BRC1 is downstream of the MORE AXILLARY GROWTH pathway and that it is required for auxin-induced apical dominance. Therefore, BRC1 acts inside the buds as an integrator of signals controlling bud outgrowth and translates them into a response of cell growth arrest. The conservation of BRC1/tb1 function among distantly related angiosperm species suggests that a single ancestral mechanism of branching control integration evolved before the radiation of flowering plants.
Figures



References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Ausin, I., Alonso-Blanco, C., and Martinez-Zapater, J.M. (2005). Environmental regulation of flowering. Int. J. Dev. Biol. 49 689–705. - PubMed
-
- Bennett, T., Sieberer, T., Willett, B., Booker, J., Luschnig, C., and Leyser, O. (2006). The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16 553–563. - PubMed
-
- Beveridge, C.A. (2006). Axillary bud outgrowth: Sending a message. Curr. Opin. Plant Biol. 9 35–40. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

