Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae
- PMID: 17308034
- PMCID: PMC1899933
- DOI: 10.1128/MCB.01084-06
Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae
Abstract
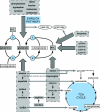
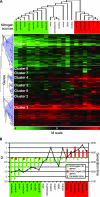
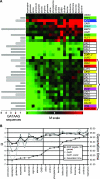
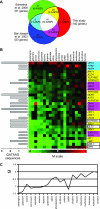
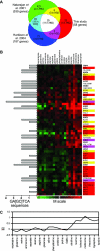
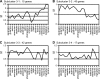
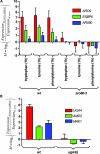
We compared the transcriptomes of Saccharomyces cerevisiae cells growing under steady-state conditions on 21 unique sources of nitrogen. We found 506 genes differentially regulated by nitrogen and estimated the activation degrees of all identified nitrogen-responding transcriptional controls according to the nitrogen source. One main group of nitrogenous compounds supports fast growth and a highly active nitrogen catabolite repression (NCR) control. Catabolism of these compounds typically yields carbon derivatives directly assimilable by a cell's metabolism. Another group of nitrogen compounds supports slower growth, is associated with excretion by cells of nonmetabolizable carbon compounds such as fusel oils, and is characterized by activation of the general control of amino acid biosynthesis (GAAC). Furthermore, NCR and GAAC appear interlinked, since expression of the GCN4 gene encoding the transcription factor that mediates GAAC is subject to NCR. We also observed that several transcriptional-regulation systems are active under a wider range of nitrogen supply conditions than anticipated. Other transcriptional-regulation systems acting on genes not involved in nitrogen metabolism, e.g., the pleiotropic-drug resistance and the unfolded-protein response systems, also respond to nitrogen. We have completed the lists of target genes of several nitrogen-sensitive regulons and have used sequence comparison tools to propose functions for about 20 orphan genes. Similar studies conducted for other nutrients should provide a more complete view of alternative metabolic pathways in yeast and contribute to the attribution of functions to many other orphan genes.
Figures


References
-
- Andre, B. 1990. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol. Gen. Genet. 220:269-276. - PubMed
-
- Andre, B., C. Hein, M. Grenson, and J. C. Jauniaux. 1993. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol. Gen. Genet. 237:17-25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
