Gene expression and biochemical analysis of cheese-ripening yeasts: focus on catabolism of L-methionine, lactate, and lactose
- PMID: 17308183
- PMCID: PMC1855621
- DOI: 10.1128/AEM.02720-06
Gene expression and biochemical analysis of cheese-ripening yeasts: focus on catabolism of L-methionine, lactate, and lactose
Abstract
DNA microarrays of 86 genes from the yeasts Debaryomyces hansenii, Kluyveromyces marxianus, and Yarrowia lipolytica were developed to determine which genes were expressed in a medium mimicking a cheese-ripening environment. These genes were selected for potential involvement in lactose/lactate catabolism and the biosynthesis of sulfur-flavored compounds. Hybridization conditions to follow specifically the expression of homologous genes belonging to different species were set up. The microarray was first validated on pure cultures of each yeast; no interspecies cross-hybridization was observed. Expression patterns of targeted genes were studied in pure cultures of each yeast, as well as in coculture, and compared to biochemical data. As expected, a high expression of the LAC genes of K. marxianus was observed. This is a yeast that efficiently degrades lactose. Several lactate dehydrogenase-encoding genes were also expressed essentially in D. hansenii and K. marxianus, which are two efficient deacidifying yeasts in cheese ripening. A set of genes possibly involved in l-methionine catabolism was also used on the array. Y. lipolytica, which efficiently assimilates l-methionine, also exhibited a high expression of the Saccharomyces cerevisiae orthologs BAT2 and ARO8, which are involved in the l-methionine degradation pathway. Our data provide the first evidence that the use of a multispecies microarray could be a powerful tool to investigate targeted metabolism and possible metabolic interactions between species within microbial cocultures.
Figures
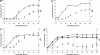
 pH levels. Error bars represent standard deviations calculated on the average values of six determinations.
pH levels. Error bars represent standard deviations calculated on the average values of six determinations.
References
-
- Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. - PubMed
-
- Arfi, K., H. E. Spinnler, R. Tâche, and P. Bonnarme. 2002. Production of volatile compounds by cheese-ripening yeasts: requirement for a methanethiol donor for S-methyl thioacetate synthesis by Kluyveromyces lactis. Appl. Microbiol. Biotechnol. 58:503-510. - PubMed
-
- Arfi, K., M. N. Leclercq-Perlat, H. E. Spinnler, and P. Bonnarme. 2005. Importance of curd-neutralising yeasts on the aromatic potential of Brevibacterium linens during cheese ripening. Int. Dairy J. 15:883-891.
-
- Barnett, J. A., R. W. Payne, and D. Yarrow. 2000. Yeasts: characteristics and identification, 3rd ed. Cambridge University Press, Cambridge, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

