Patterns and rates of intron divergence between humans and chimpanzees
- PMID: 17309804
- PMCID: PMC1852421
- DOI: 10.1186/gb-2007-8-2-r21
Patterns and rates of intron divergence between humans and chimpanzees
Abstract
Background: Introns, which constitute the largest fraction of eukaryotic genes and which had been considered to be neutral sequences, are increasingly acknowledged as having important functions. Several studies have investigated levels of evolutionary constraint along introns and across classes of introns of different length and location within genes. However, thus far these studies have yielded contradictory results.
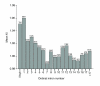
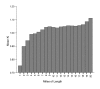
Results: We present the first analysis of human-chimpanzee intron divergence, in which differences in the number of substitutions per intronic site (Ki) can be interpreted as the footprint of different intensities and directions of the pressures of natural selection. Our main findings are as follows: there was a strong positive correlation between intron length and divergence; there was a strong negative correlation between intron length and GC content; and divergence rates vary along introns and depending on their ordinal position within genes (for instance, first introns are more GC rich, longer and more divergent, and divergence is lower at the 3' and 5' ends of all types of introns).
Conclusion: We show that the higher divergence of first introns is related to their larger size. Also, the lower divergence of short introns suggests that they may harbor a relatively greater proportion of regulatory elements than long introns. Moreover, our results are consistent with the presence of functionally relevant sequences near the 5' and 3' ends of introns. Finally, our findings suggest that other parts of introns may also be under selective constraints.
Figures
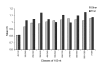
Similar articles
-
Analysis of Five Gene Sets in Chimpanzees Suggests Decoupling between the Action of Selection on Protein-Coding and on Noncoding Elements.Genome Biol Evol. 2015 May 14;7(6):1490-505. doi: 10.1093/gbe/evv082. Genome Biol Evol. 2015. PMID: 25977458 Free PMC article.
-
Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content.Genome Biol. 2005;6(8):R67. doi: 10.1186/gb-2005-6-8-r67. Epub 2005 Jul 27. Genome Biol. 2005. PMID: 16086849 Free PMC article.
-
Patterns of exon-intron architecture variation of genes in eukaryotic genomes.BMC Genomics. 2009 Jan 24;10:47. doi: 10.1186/1471-2164-10-47. BMC Genomics. 2009. PMID: 19166620 Free PMC article.
-
Intron-rich ancestors.Trends Genet. 2006 Sep;22(9):468-71. doi: 10.1016/j.tig.2006.07.002. Epub 2006 Jul 20. Trends Genet. 2006. PMID: 16857287 Review.
-
Recombination rate variation in closely related species.Heredity (Edinb). 2011 Dec;107(6):496-508. doi: 10.1038/hdy.2011.44. Epub 2011 Jun 15. Heredity (Edinb). 2011. PMID: 21673743 Free PMC article. Review.
Cited by
-
In with the old, in with the new: the promiscuity of the duplication process engenders diverse pathways for novel gene creation.Int J Evol Biol. 2012;2012:341932. doi: 10.1155/2012/341932. Epub 2012 Sep 13. Int J Evol Biol. 2012. PMID: 23008799 Free PMC article.
-
Analysis of Five Gene Sets in Chimpanzees Suggests Decoupling between the Action of Selection on Protein-Coding and on Noncoding Elements.Genome Biol Evol. 2015 May 14;7(6):1490-505. doi: 10.1093/gbe/evv082. Genome Biol Evol. 2015. PMID: 25977458 Free PMC article.
-
Genome-wide evolutionary analysis of the noncoding RNA genes and noncoding DNA of Paramecium tetraurelia.RNA. 2009 Apr;15(4):503-14. doi: 10.1261/rna.1306009. Epub 2009 Feb 13. RNA. 2009. PMID: 19218550 Free PMC article.
-
Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce.Mol Ecol. 2008 Aug;17(16):3599-613. doi: 10.1111/j.1365-294X.2008.03840.x. Epub 2008 Jul 3. Mol Ecol. 2008. PMID: 18662225 Free PMC article.
-
Intragenomic conflict associated with extreme phenotypic plasticity in queen-worker caste determination in honey bees (Apis mellifera).Genome Biol. 2025 Jun 18;26(1):171. doi: 10.1186/s13059-025-03628-0. Genome Biol. 2025. PMID: 40533789 Free PMC article.
References
-
- Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

