Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA
- PMID: 17311413
- PMCID: PMC2271177
- DOI: 10.1021/bi0619472
Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA
Abstract
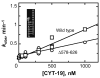
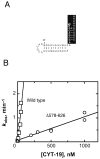
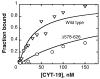
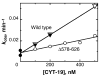
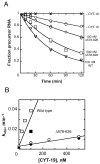
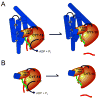
The DEAD-box protein CYT-19 functions in the folding of several group I introns in vivo and a diverse set of group I and group II RNAs in vitro. Recent work using the Tetrahymena group I ribozyme demonstrated that CYT-19 possesses a second RNA-binding site, distinct from the unwinding active site, which enhances unwinding activity by binding nonspecifically to the adjacent RNA structure. Here, we probe the region of CYT-19 responsible for that binding by constructing a C-terminal truncation variant that lacks 49 amino acids and terminates at a domain boundary, as defined by limited proteolysis. This truncated protein unwinds a six-base-pair duplex, formed between the oligonucleotide substrate of the Tetrahymena ribozyme and an oligonucleotide corresponding to the internal guide sequence of the ribozyme, with near-wild-type efficiency. However, the truncated protein is activated much less than the wild-type protein when the duplex is covalently linked to the ribozyme or single-stranded or double-stranded extensions. Thus, the active site for RNA unwinding remains functional in the truncated CYT-19, but the site that binds the adjacent RNA structure has been compromised. Equilibrium binding experiments confirmed that the truncated protein binds RNA less tightly than the wild-type protein. RNA binding by the compromised site is important for chaperone activity, because the truncated protein is less active in facilitating the folding of a group I intron that requires CYT-19 in vivo. The deleted region contains arginine-rich sequences, as found in other RNA-binding proteins, and may function by tethering CYT-19 to structured RNAs, so that it can efficiently disrupt exposed, non-native structural elements, allowing them to refold. Many other DExD/H-box proteins also contain arginine-rich ancillary domains, and some of these domains may function similarly as nonspecific RNA-binding elements that enhance general RNA chaperone activity.
Figures


Similar articles
-
DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA.Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):E2928-36. doi: 10.1073/pnas.1404307111. Epub 2014 Jul 7. Proc Natl Acad Sci U S A. 2014. PMID: 25002474 Free PMC article.
-
Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone.Proc Natl Acad Sci U S A. 2006 Nov 7;103(45):16698-703. doi: 10.1073/pnas.0603127103. Epub 2006 Oct 30. Proc Natl Acad Sci U S A. 2006. PMID: 17075070 Free PMC article.
-
Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity.J Mol Biol. 2007 Jan 19;365(3):835-55. doi: 10.1016/j.jmb.2006.09.083. Epub 2006 Oct 3. J Mol Biol. 2007. PMID: 17081564 Free PMC article.
-
DEAD-box proteins as RNA helicases and chaperones.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):135-52. doi: 10.1002/wrna.50. Wiley Interdiscip Rev RNA. 2011. PMID: 21297876 Free PMC article. Review.
-
Iterative annealing mechanism explains the functions of the GroEL and RNA chaperones.Protein Sci. 2020 Feb;29(2):360-377. doi: 10.1002/pro.3795. Epub 2019 Dec 23. Protein Sci. 2020. PMID: 31800116 Free PMC article. Review.
Cited by
-
Toward a molecular understanding of RNA remodeling by DEAD-box proteins.RNA Biol. 2013 Jan;10(1):44-55. doi: 10.4161/rna.22210. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995827 Free PMC article. Review.
-
DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA.Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):E2928-36. doi: 10.1073/pnas.1404307111. Epub 2014 Jul 7. Proc Natl Acad Sci U S A. 2014. PMID: 25002474 Free PMC article.
-
DEAD-box proteins can completely separate an RNA duplex using a single ATP.Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20203-8. doi: 10.1073/pnas.0811075106. Epub 2008 Dec 16. Proc Natl Acad Sci U S A. 2008. PMID: 19088196 Free PMC article.
-
Role of RNA chaperones in virus replication.Virus Res. 2009 Feb;139(2):253-66. doi: 10.1016/j.virusres.2008.06.015. Epub 2008 Aug 8. Virus Res. 2009. PMID: 18675859 Free PMC article. Review.
-
The DEAD-Box Protein CYT-19 Uses Arginine Residues in Its C-Tail To Tether RNA Substrates.Biochemistry. 2017 Jul 18;56(28):3571-3578. doi: 10.1021/acs.biochem.7b00362. Epub 2017 Jul 7. Biochemistry. 2017. PMID: 28650145 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

