Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids
- PMID: 17311473
- PMCID: PMC1800307
- DOI: 10.1371/journal.pbio.0050059
Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids
Abstract
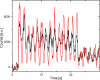
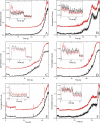
The bacteriophage phi29 generates large forces to compact its double-stranded DNA genome into a protein capsid by means of a portal motor complex. Several mechanical models for the generation of these high forces by the motor complex predict coupling of DNA translocation to rotation of the head-tail connector dodecamer. Putative connector rotation is investigated here by combining the methods of single-molecule force spectroscopy with polarization-sensitive single-molecule fluorescence. In our experiment, we observe motor function in several packaging complexes in parallel using video microscopy of bead position in a magnetic trap. At the same time, we follow the orientation of single fluorophores attached to the portal motor connector. From our data, we can exclude connector rotation with greater than 99% probability and therefore answer a long-standing mechanistic question.
Conflict of interest statement
Figures
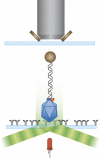
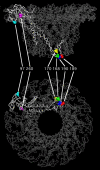
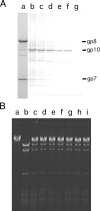
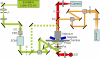
Comment in
-
Does bacteriophage phi29 pack its DNA with a twist?PLoS Biol. 2007 Mar;5(3):e91. doi: 10.1371/journal.pbio.0050091. Epub 2007 Feb 20. PLoS Biol. 2007. PMID: 20076665 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

