Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression
- PMID: 17312117
- PMCID: PMC4520734
- DOI: 10.4049/jimmunol.178.5.2746
Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression
Abstract
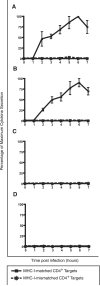
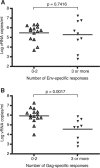
CD8(+) T cells are a key focus of vaccine development efforts for HIV. However, there is no clear consensus as to which of the nine HIV proteins should be used for vaccination. The early proteins Tat, Rev, and Nef may be better CD8(+) T cell targets than the late-expressed structural proteins Gag, Pol, and Env. In this study, we show that Gag-specific CD8(+) T cells recognize infected CD4(+) T lymphocytes as early as 2 h postinfection, before proviral DNA integration, viral protein synthesis, and Nef-mediated MHC class I down-regulation. Additionally, the number of Gag epitopes recognized by CD8(+) T cells was significantly associated with lower viremia (p = 0.0017) in SIV-infected rhesus macaques. These results suggest that HIV vaccines should focus CD8(+) T cell responses on Gag.
Figures





Similar articles
-
Containment of simian immunodeficiency virus infection in vaccinated macaques: correlation with the magnitude of virus-specific pre- and postchallenge CD4+ and CD8+ T cell responses.J Immunol. 2002 Nov 1;169(9):4778-87. doi: 10.4049/jimmunol.169.9.4778. J Immunol. 2002. PMID: 12391187
-
Utility of human immunodeficiency virus type 1 envelope as a T-cell immunogen.J Virol. 2007 Dec;81(23):13125-34. doi: 10.1128/JVI.01408-07. Epub 2007 Sep 26. J Virol. 2007. PMID: 17898063 Free PMC article.
-
The Frequency of Vaccine-Induced T-Cell Responses Does Not Predict the Rate of Acquisition after Repeated Intrarectal SIVmac239 Challenges in Mamu-B*08+ Rhesus Macaques.J Virol. 2019 Feb 19;93(5):e01626-18. doi: 10.1128/JVI.01626-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541854 Free PMC article.
-
A replication competent adenovirus 5 host range mutant-simian immunodeficiency virus (SIV) recombinant priming/subunit protein boosting vaccine regimen induces broad, persistent SIV-specific cellular immunity to dominant and subdominant epitopes in Mamu-A*01 rhesus macaques.J Immunol. 2003 Apr 15;170(8):4281-9. doi: 10.4049/jimmunol.170.8.4281. J Immunol. 2003. PMID: 12682263
-
The hope for an HIV vaccine based on induction of CD8+ T lymphocytes--a review.Mem Inst Oswaldo Cruz. 2008 Mar;103(2):119-29. doi: 10.1590/s0074-02762008000200001. Mem Inst Oswaldo Cruz. 2008. PMID: 18425263 Free PMC article. Review.
Cited by
-
Anti-HIV Antibody Responses and the HIV Reservoir Size during Antiretroviral Therapy.PLoS One. 2016 Aug 2;11(8):e0160192. doi: 10.1371/journal.pone.0160192. eCollection 2016. PLoS One. 2016. PMID: 27483366 Free PMC article.
-
A novel recombinant bacterial vaccine strain expressing dual viral antigens induces multiple immune responses to the Gag and gp120 proteins of HIV-1 in immunized mice.Antiviral Res. 2008 Dec;80(3):272-9. doi: 10.1016/j.antiviral.2008.06.013. Epub 2008 Jul 17. Antiviral Res. 2008. PMID: 18639586 Free PMC article.
-
For protection from HIV-1 infection, more might not be better: a systematic analysis of HIV Gag epitopes of two alleles associated with different outcomes of HIV-1 infection.J Virol. 2012 Jan;86(2):1166-80. doi: 10.1128/JVI.05721-11. Epub 2011 Nov 9. J Virol. 2012. PMID: 22072744 Free PMC article.
-
Magnitude and complexity of rectal mucosa HIV-1-specific CD8+ T-cell responses during chronic infection reflect clinical status.PLoS One. 2008;3(10):e3577. doi: 10.1371/journal.pone.0003577. Epub 2008 Oct 30. PLoS One. 2008. PMID: 18974782 Free PMC article.
-
Implications of CTL-mediated killing of HIV-infected cells during the non-productive stage of infection.PLoS One. 2011 Feb 7;6(2):e16468. doi: 10.1371/journal.pone.0016468. PLoS One. 2011. PMID: 21326882 Free PMC article.
References
-
- Baba TW, Liska V, Khimani AH, Ray NB, Dailey PJ, Penninck D, Bronson R, Greene MF, McClure HM, Martin LN, Ruprecht RM. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 1999;5:194–203. - PubMed
-
- VaxGen . VaxGen Announces Results of its Phase III HIV Vaccine Trial in Thailand: Vaccine Fails to Meet Endpoints. VaxGen; Brisbane: 2003.
-
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 2004;5:233–236. - PubMed
-
- Wilson NA, Reed J, Napoe GS, Piaskowski S, Szymanski A, Furlott J, Gonzalez EJ, Yant LJ, Maness NJ, May GE, et al. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 2006;80:5875–5885. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI052056/AI/NIAID NIH HHS/United States
- R37 AI052056/AI/NIAID NIH HHS/United States
- RR 15459-01/RR/NCRR NIH HHS/United States
- P51 RR 000167/RR/NCRR NIH HHS/United States
- RR 020141-01/RR/NCRR NIH HHS/United States
- R01 AI 049120/AI/NIAID NIH HHS/United States
- N01 CO012400/CA/NCI NIH HHS/United States
- R01 AI049120/AI/NIAID NIH HHS/United States
- R01 AI 052056/AI/NIAID NIH HHS/United States
- N01 CO012400/CO/NCI NIH HHS/United States
- C06 RR020141/RR/NCRR NIH HHS/United States
- C06 RR015459/RR/NCRR NIH HHS/United States
- P51 RR000167/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

