Live covisualization of competing adeno-associated virus and herpes simplex virus type 1 DNA replication: molecular mechanisms of interaction
- PMID: 17314170
- PMCID: PMC1900138
- DOI: 10.1128/JVI.02476-06
Live covisualization of competing adeno-associated virus and herpes simplex virus type 1 DNA replication: molecular mechanisms of interaction
Abstract
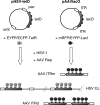
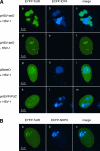
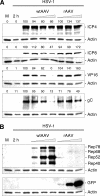
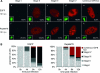
We performed live cell visualization assays to directly assess the interaction between competing adeno-associated virus (AAV) and herpes simplex virus type 1 (HSV-1) DNA replication. Our studies reveal the formation of separate AAV and HSV-1 replication compartments and the inhibition of HSV-1 replication compartment formation in the presence of AAV. AAV Rep is recruited into AAV replication compartments but not into those of HSV-1, while the single-stranded DNA-binding protein HSV-1 ICP8 is recruited into both AAV and HSV-1 replication compartments, although with differential staining patterns. Slot blot analysis of coinfected cells revealed a dose-dependent inhibition of HSV-1 DNA replication by wild-type AAV but not by rep-negative recombinant AAV. Consistent with this, Western blot analysis indicated that wild-type AAV affects the levels of the HSV-1 immediate-early protein ICP4 and the early protein ICP8 only modestly but strongly inhibits the accumulation of the late proteins VP16 and gC. Furthermore, we demonstrate that the presence of Rep in the absence of AAV DNA replication is sufficient for the inhibition of HSV-1. In particular, Rep68/78 proteins severely inhibit the formation of mature HSV-1 replication compartments and lead to the accumulation of ICP8 at sites of cellular DNA synthesis, a phenomenon previously observed in the presence of viral polymerase inhibitors. Taken together, our results suggest that AAV and HSV-1 replicate in separate compartments and that AAV Rep inhibits HSV-1 at the level of DNA replication.
Figures



Similar articles
-
Identification of rep-associated factors in herpes simplex virus type 1-induced adeno-associated virus type 2 replication compartments.J Virol. 2010 Sep;84(17):8871-87. doi: 10.1128/JVI.00725-10. Epub 2010 Jun 23. J Virol. 2010. PMID: 20573815 Free PMC article.
-
Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events.PLoS Pathog. 2009 Mar;5(3):e1000340. doi: 10.1371/journal.ppat.1000340. Epub 2009 Mar 13. PLoS Pathog. 2009. PMID: 19282980 Free PMC article.
-
The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication.J Virol. 2004 Jan;78(1):441-53. doi: 10.1128/jvi.78.1.441-453.2004. J Virol. 2004. PMID: 14671124 Free PMC article.
-
Chimeric herpes simplex virus/adeno-associated virus amplicon vectors.Curr Gene Ther. 2006 Jun;6(3):315-24. doi: 10.2174/156652306777592090. Curr Gene Ther. 2006. PMID: 16787183 Review.
-
Herpes simplex virus type-1: a model for genome transactions.Prog Nucleic Acid Res Mol Biol. 2003;75:139-71. doi: 10.1016/s0079-6603(03)75005-3. Prog Nucleic Acid Res Mol Biol. 2003. PMID: 14604012 Review.
Cited by
-
Initiation of lytic DNA replication in Epstein-Barr virus: search for a common family mechanism.Future Virol. 2010 Jan;5(1):65-83. doi: 10.2217/fvl.09.69. Future Virol. 2010. PMID: 22468146 Free PMC article.
-
Identification of rep-associated factors in herpes simplex virus type 1-induced adeno-associated virus type 2 replication compartments.J Virol. 2010 Sep;84(17):8871-87. doi: 10.1128/JVI.00725-10. Epub 2010 Jun 23. J Virol. 2010. PMID: 20573815 Free PMC article.
-
Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase.J Virol. 2009 Jun;83(12):6269-78. doi: 10.1128/JVI.00318-09. Epub 2009 Apr 1. J Virol. 2009. PMID: 19339345 Free PMC article.
-
Large-scale adeno-associated viral vector production using a herpesvirus-based system enables manufacturing for clinical studies.Hum Gene Ther. 2009 Aug;20(8):796-806. doi: 10.1089/hum.2009.094. Hum Gene Ther. 2009. PMID: 19569968 Free PMC article. Review.
-
Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events.PLoS Pathog. 2009 Mar;5(3):e1000340. doi: 10.1371/journal.ppat.1000340. Epub 2009 Mar 13. PLoS Pathog. 2009. PMID: 19282980 Free PMC article.
References
-
- Bantel-Schaal, U., and H. zur Hausen. 1988. Adeno-associated viruses inhibit SV40 DNA amplification and replication of herpes simplex virus in SV40-transformed hamster cells. Virology 164:64-74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

