Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase
- PMID: 17316440
- PMCID: PMC1852407
- DOI: 10.1186/gb-2007-8-2-r23
Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase
Abstract
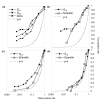
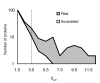
Protein kinases are critical to cellular signalling and post-translational gene regulation, but their biological substrates are difficult to identify. We show that cyclin-dependent kinase (CDK) consensus motifs are frequently clustered in CDK substrate proteins. Based on this, we introduce a new computational strategy to predict the targets of CDKs and use it to identify new biologically interesting candidates. Our data suggest that regulatory modules may exist in protein sequence as clusters of short sequence motifs.
Figures

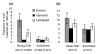



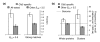
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

