INO80 subfamily of chromatin remodeling complexes
- PMID: 17316710
- PMCID: PMC2699258
- DOI: 10.1016/j.mrfmmm.2006.10.006
INO80 subfamily of chromatin remodeling complexes
Abstract
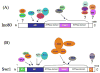
ATP-dependent chromatin remodeling complexes contain ATPases of the Swi/Snf superfamily and alter DNA accessibility of chromatin in an ATP-dependent manner. Recently characterized INO80 and SWR1 complexes belong to a subfamily of these chromatin remodelers and are characterized by a split ATPase domain in the core ATPase subunit and the presence of Rvb proteins. INO80 and SWR1 complexes are evolutionarily conserved from yeast to human and have been implicated in transcription regulation, as well as DNA repair. The individual components, assembly patterns, and molecular mechanisms of the INO80 class of chromatin remodeling complexes are discussed in this review.
Figures
References
-
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr Opin Genet Dev. 2004;14:165–173. - PubMed
-
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev. 2001;11:148–154. - PubMed
-
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. - PubMed
-
- Muchardt C, Yaniv M. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J Mol Biol. 1999;293:187–198. - PubMed
-
- Peterson CL. ATP-dependent chromatin remodeling: going mobile. FEBS Lett. 2000;476:68–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

