A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function
- PMID: 17317571
- PMCID: PMC1858662
- DOI: 10.1016/j.chembiol.2006.12.008
A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function
Abstract
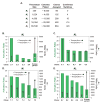
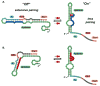
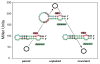
Riboswitches are RNA-based genetic control elements that regulate gene expression in a ligand-dependent fashion without the need for proteins. The ability to create synthetic riboswitches that control gene expression in response to any desired small-molecule ligand will enable the development of sensitive genetic screens that can detect the presence of small molecules, as well as designer genetic control elements to conditionally modulate cellular behavior. Herein, we present an automated high-throughput screening method that identifies synthetic riboswitches that display extremely low background levels of gene expression in the absence of the desired ligand and robust increases in expression in its presence. Mechanistic studies reveal how these riboswitches function and suggest design principles for creating new synthetic riboswitches. We anticipate that the screening method and design principles will be generally useful for creating functional synthetic riboswitches.
Figures
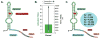


Similar articles
-
Riboswitches in unexpected places--a synthetic riboswitch in a protein coding region.RNA. 2008 Dec;14(12):2498-503. doi: 10.1261/rna.1269008. Epub 2008 Oct 22. RNA. 2008. PMID: 18945803 Free PMC article.
-
Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation.J Am Chem Soc. 2004 Oct 20;126(41):13247-54. doi: 10.1021/ja048634j. J Am Chem Soc. 2004. PMID: 15479078
-
A flow cytometry-based screen for synthetic riboswitches.Nucleic Acids Res. 2009 Jan;37(1):184-92. doi: 10.1093/nar/gkn924. Epub 2008 Nov 25. Nucleic Acids Res. 2009. PMID: 19033367 Free PMC article.
-
Riboswitches as antibacterial drug targets.Nat Biotechnol. 2006 Dec;24(12):1558-64. doi: 10.1038/nbt1268. Nat Biotechnol. 2006. PMID: 17160062 Review.
-
Expanding roles for metabolite-sensing regulatory RNAs.Curr Opin Microbiol. 2009 Apr;12(2):161-9. doi: 10.1016/j.mib.2009.01.012. Epub 2009 Feb 26. Curr Opin Microbiol. 2009. PMID: 19250859 Free PMC article. Review.
Cited by
-
SHAPE-directed RNA secondary structure prediction.Methods. 2010 Oct;52(2):150-8. doi: 10.1016/j.ymeth.2010.06.007. Epub 2010 Jun 8. Methods. 2010. PMID: 20554050 Free PMC article. Review.
-
Riboswitches in unexpected places--a synthetic riboswitch in a protein coding region.RNA. 2008 Dec;14(12):2498-503. doi: 10.1261/rna.1269008. Epub 2008 Oct 22. RNA. 2008. PMID: 18945803 Free PMC article.
-
A family of synthetic riboswitches adopts a kinetic trapping mechanism.Nucleic Acids Res. 2014 Jun;42(10):6753-61. doi: 10.1093/nar/gku262. Epub 2014 Apr 29. Nucleic Acids Res. 2014. PMID: 24782524 Free PMC article.
-
Evaluating riboswitch optimality.Methods Enzymol. 2019;623:417-450. doi: 10.1016/bs.mie.2019.05.028. Epub 2019 Jun 18. Methods Enzymol. 2019. PMID: 31239056 Free PMC article.
-
Design of Artificial Riboswitches as Biosensors.Sensors (Basel). 2017 Aug 30;17(9):1990. doi: 10.3390/s17091990. Sensors (Basel). 2017. PMID: 28867802 Free PMC article. Review.
References
-
- Winkler WC, Breaker RR. REGULATION OF BACTERIAL GENE EXPRESSION BY RIBOSWITCHES. Annual Review of Microbiology. 2005;59:487–517. - PubMed
-
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. - PubMed
-
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 2004;20:44–50. - PubMed
-
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Current Opinion in Structural Biology. 2005;15:342. - PubMed
-
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

