Identification of two distinct hybrid state intermediates on the ribosome
- PMID: 17317624
- PMCID: PMC2244649
- DOI: 10.1016/j.molcel.2007.01.022
Identification of two distinct hybrid state intermediates on the ribosome
Abstract
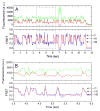
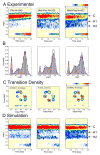
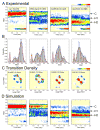
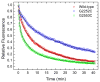
High spatial and time resolution single-molecule fluorescence resonance energy transfer measurements have been used to probe the structural and kinetic parameters of transfer RNA (tRNA) movements within the aminoacyl (A) and peptidyl (P) sites of the ribosome. Our investigation of tRNA motions, quantified on wild-type, mutant, and L1-depleted ribosome complexes, reveals a dynamic exchange between three metastable tRNA configurations, one of which is a previously unidentified hybrid state in which only deacylated-tRNA adopts its hybrid (P/E) configuration. This new dynamic information suggests a framework in which the formation of intermediate states in the translocation process is achieved through global conformational rearrangements of the ribosome particle.
Figures
References
-
- Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J. Effect of Buffer Conditions on the Position of tRNA on the 70S Ribosome As Visualized by Cryoelectron Microscopy. J Biol Chem. 1999a;274:8723–8729. - PubMed
-
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol. 1999b;6:643–647. - PubMed
-
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004b;11:1008–1014. - PubMed
-
- Blaha G, Burkhardt N, Nierhaus K. Formation of 70S ribosomes: large activation energy is required for the adaptation of exclusively the small ribosomal subunit. Biophys Chem. 2002;96:153–161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

