Phosphorylation of HuR by Chk2 regulates SIRT1 expression
- PMID: 17317627
- PMCID: PMC1986740
- DOI: 10.1016/j.molcel.2007.01.011
Phosphorylation of HuR by Chk2 regulates SIRT1 expression
Abstract
The RNA binding protein HuR regulates the stability of many target mRNAs. Here, we report that HuR associated with the 3' untranslated region of the mRNA encoding the longevity and stress-response protein SIRT1, stabilized the SIRT1 mRNA, and increased SIRT1 expression levels. Unexpectedly, oxidative stress triggered the dissociation of the [HuR-SIRT1 mRNA] complex, in turn promoting SIRT1 mRNA decay, reducing SIRT1 abundance, and lowering cell survival. The cell cycle checkpoint kinase Chk2 was activated by H(2)O(2), interacted with HuR, and was predicted to phosphorylate HuR at residues S88, S100, and T118. Mutation of these residues revealed a complex pattern of HuR binding, with S100 appearing to be important for [HuR-SIRT1 mRNA] dissociation after H(2)O(2). Our findings demonstrate that HuR regulates SIRT1 expression, underscore functional links between the two stress-response proteins, and implicate Chk2 in these processes.
Figures

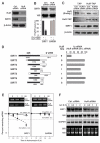

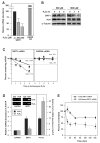

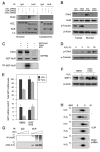
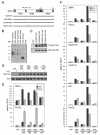
Comment in
-
HuR-SIRT: the hairy world of posttranscriptional control.Mol Cell. 2007 Feb 23;25(4):485-7. doi: 10.1016/j.molcel.2007.02.001. Mol Cell. 2007. PMID: 17317621
References
-
- Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. - PubMed
-
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. - PubMed
-
- Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell Physiol. 2003;195:356–372. - PubMed
-
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

