Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector
- PMID: 17317684
- PMCID: PMC1874629
- DOI: 10.1093/nar/gkm078
Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector
Abstract
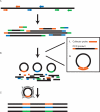
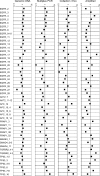
Herein we present Gene-Collector, a method for multiplex amplification of nucleic acids. The procedure has been employed to successfully amplify the coding sequence of 10 human cancer genes in one assay with uniform abundance of the final products. Amplification is initiated by a multiplex PCR in this case with 170 primer pairs. Each PCR product is then specifically circularized by ligation on a Collector probe capable of juxtapositioning only the perfectly matched cognate primer pairs. Any amplification artifacts typically associated with multiplex PCR derived from the use of many primer pairs such as false amplicons, primer-dimers etc. are not circularized and degraded by exonuclease treatment. Circular DNA molecules are then further enriched by randomly primed rolling circle replication. Amplification was successful for 90% of the targeted amplicons as seen by hybridization to a custom resequencing DNA micro-array. Real-time quantitative PCR revealed that 96% of the amplification products were all within 4-fold of the average abundance. Gene-Collector has utility for numerous applications such as high throughput resequencing, SNP analyses, and pathogen detection.
Figures
References
-
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, et al. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. - PubMed
-
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. - PubMed
-
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005 1117389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

