A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay
- PMID: 17318186
- PMCID: PMC1829367
- DOI: 10.1038/sj.emboj.7601588
A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay
Abstract
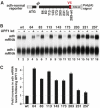
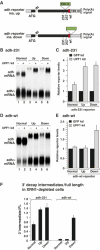
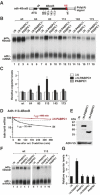
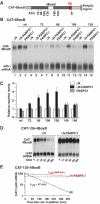
The nonsense-mediated mRNA decay (NMD) pathway degrades mRNAs with premature translation termination codons (PTCs). The mechanisms by which PTCs and natural stop codons are discriminated remain unclear. We show that the position of stops relative to the poly(A) tail (and thus of PABPC1) is a critical determinant for PTC definition in Drosophila melanogaster. Indeed, tethering of PABPC1 downstream of a PTC abolishes NMD. Conversely, natural stops trigger NMD when the length of the 3' UTR is increased. However, many endogenous transcripts with exceptionally long 3' UTRs escape NMD, suggesting that the increase in 3' UTR length has co-evolved with the acquisition of features that suppress NMD. We provide evidence for the existence of 3' UTRs conferring immunity to NMD. We also show that PABPC1 binding is sufficient for PTC recognition, regardless of cleavage or polyadenylation. The role of PABPC1 in NMD must go beyond that of providing positional information for PTC definition, because its depletion suppresses NMD under conditions in which translation efficiency is not affected. These findings reveal a conserved role for PABPC1 in mRNA surveillance.
Figures


References
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
-
- Amrani N, Sachs MS, Jacobson A (2006) Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7: 415–425 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

