A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans
- PMID: 17319742
- PMCID: PMC1803011
- DOI: 10.1371/journal.ppat.0030022
A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans
Abstract
Fungal pathogens of humans require molecular oxygen for several essential biochemical reactions, yet virtually nothing is known about how they adapt to the relatively hypoxic environment of infected tissues. We isolated mutants defective in growth under hypoxic conditions, but normal for growth in normoxic conditions, in Cryptococcus neoformans, the most common cause of fungal meningitis. Two regulatory pathways were identified: one homologous to the mammalian sterol-response element binding protein (SREBP) cholesterol biosynthesis regulatory pathway, and the other a two-component-like pathway involving a fungal-specific hybrid histidine kinase family member, Tco1. We show that cleavage of the SREBP precursor homolog Sre1-which is predicted to release its DNA-binding domain from the membrane-occurs in response to hypoxia, and that Sre1 is required for hypoxic induction of genes encoding for oxygen-dependent enzymes involved in ergosterol synthesis. Importantly, mutants in either the SREBP pathway or the Tco1 pathway display defects in their ability to proliferate in host tissues and to cause disease in infected mice, linking for the first time to our knowledge hypoxic adaptation and pathogenesis by a eukaryotic aerobe. SREBP pathway mutants were found to be a hundred times more sensitive than wild-type to fluconazole, a widely used antifungal agent that inhibits ergosterol synthesis, suggesting that inhibitors of SREBP processing could substantially enhance the potency of current therapies.
Conflict of interest statement
Figures


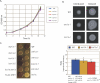
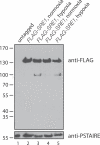
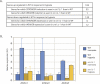

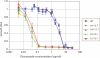
References
-
- Erecinska M, Silver IA. Tissue oxygen tension and brain sensitivity to hypoxia. Respir Physiol. 2001;128:263–276. - PubMed
-
- Carlsson PO, Palm F, Andersson A, Liss P. Markedly decreased oxygen tension in transplanted rat pancreatic islets irrespective of the implantation site. Diabetes. 2001;50:489–495. - PubMed
-
- Nau R, Bruck W. Neuronal injury in bacterial meningitis: Mechanisms and implications for therapy. Trends Neurosci. 2002;25:38–45. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

