Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model
- PMID: 17319744
- PMCID: PMC1802818
- DOI: 10.1371/journal.pgen.0030007
Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model
Abstract
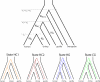
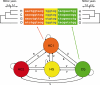
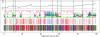
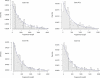
The genealogical relationship of human, chimpanzee, and gorilla varies along the genome. We develop a hidden Markov model (HMM) that incorporates this variation and relate the model parameters to population genetics quantities such as speciation times and ancestral population sizes. Our HMM is an analytically tractable approximation to the coalescent process with recombination, and in simulations we see no apparent bias in the HMM estimates. We apply the HMM to four autosomal contiguous human-chimp-gorilla-orangutan alignments comprising a total of 1.9 million base pairs. We find a very recent speciation time of human-chimp (4.1 +/- 0.4 million years), and fairly large ancestral effective population sizes (65,000 +/- 30,000 for the human-chimp ancestor and 45,000 +/- 10,000 for the human-chimp-gorilla ancestor). Furthermore, around 50% of the human genome coalesces with chimpanzee after speciation with gorilla. We also consider 250,000 base pairs of X-chromosome alignments and find an effective population size much smaller than 75% of the autosomal effective population sizes. Finally, we find that the rate of transitions between different genealogies correlates well with the region-wide present-day human recombination rate, but does not correlate with the fine-scale recombination rates and recombination hot spots, suggesting that the latter are evolutionarily transient.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures




Similar articles
-
Ancestral population genomics: the coalescent hidden Markov model approach.Genetics. 2009 Sep;183(1):259-74. doi: 10.1534/genetics.109.103010. Epub 2009 Jul 6. Genetics. 2009. PMID: 19581452 Free PMC article.
-
Reconstructing the demographic history of the human lineage using whole-genome sequences from human and three great apes.Genome Biol Evol. 2012;4(11):1133-45. doi: 10.1093/gbe/evs075. Genome Biol Evol. 2012. PMID: 22975719 Free PMC article.
-
A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species.PLoS Genet. 2012;8(12):e1003125. doi: 10.1371/journal.pgen.1003125. Epub 2012 Dec 20. PLoS Genet. 2012. PMID: 23284294 Free PMC article.
-
TRAILS: Tree reconstruction of ancestry using incomplete lineage sorting.PLoS Genet. 2024 Feb 8;20(2):e1010836. doi: 10.1371/journal.pgen.1010836. eCollection 2024 Feb. PLoS Genet. 2024. PMID: 38330138 Free PMC article.
-
[Molecular phylogeny of humans, chimpanzees and gorillas].Tanpakushitsu Kakusan Koso. 2000 Dec;45(16):2588-95. Tanpakushitsu Kakusan Koso. 2000. PMID: 11185912 Review. Japanese. No abstract available.
Cited by
-
Chromosome-scale inference of hybrid speciation and admixture with convolutional neural networks.Mol Ecol Resour. 2021 Nov;21(8):2676-2688. doi: 10.1111/1755-0998.13355. Epub 2021 Mar 8. Mol Ecol Resour. 2021. PMID: 33682305 Free PMC article.
-
Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis.Genome Res. 2012 Apr;22(4):746-54. doi: 10.1101/gr.125864.111. Epub 2011 Dec 29. Genome Res. 2012. PMID: 22207614 Free PMC article.
-
Next-generation hybridization and introgression.Heredity (Edinb). 2012 Mar;108(3):179-89. doi: 10.1038/hdy.2011.68. Epub 2011 Sep 7. Heredity (Edinb). 2012. PMID: 21897439 Free PMC article. Review.
-
Genome-wide inference of ancestral recombination graphs.PLoS Genet. 2014 May 15;10(5):e1004342. doi: 10.1371/journal.pgen.1004342. eCollection 2014. PLoS Genet. 2014. PMID: 24831947 Free PMC article.
-
Methods and models for unravelling human evolutionary history.Nat Rev Genet. 2015 Dec;16(12):727-40. doi: 10.1038/nrg4005. Epub 2015 Nov 10. Nat Rev Genet. 2015. PMID: 26553329 Review.
References
-
- Enard W, Pääbo S. Comparative primate genomics. Annu Rev Genomics Hum Genet. 2004;5:351–378. - PubMed
-
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of human and chimpanzees. Nature. 2006;441:1103–1108. - PubMed
-
- Innan H, Watanabe H. The effect of gene flow on the coalescent time in the human-chimpanzee ancestral population. Mol Biol Evol. 2006;23:1040–1047. - PubMed
-
- Barton NH. Evolutionary biology: How did the human species form? Curr Biol. 2006;16:R647–R650. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

