Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins
- PMID: 17319956
- PMCID: PMC1810245
- DOI: 10.1186/1471-2148-7-29
Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins
Abstract
Background: In membrane trafficking, the mechanisms ensuring vesicle fusion specificity remain to be fully elucidated. Early models proposed that specificity was encoded entirely by SNARE proteins; more recent models include contributions from Rab proteins, Syntaxin-binding (SM) proteins and tethering factors. Most information on membrane trafficking derives from an evolutionarily narrow sampling of model organisms. However, considering factors from a wider diversity of eukaryotes can provide both functional information on core systems and insight into the evolutionary history of the trafficking machinery. For example, the major Qa/syntaxin SNARE families are present in most eukaryotic genomes and likely each evolved via gene duplication from a single ancestral syntaxin before the existing eukaryotic groups diversified. This pattern is also likely for Rabs and various other components of the membrane trafficking machinery.
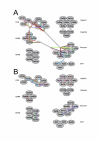
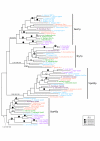
Results: We performed comparative genomic and phylogenetic analyses, when relevant, on the SM proteins and components of the tethering complexes, both thought to contribute to vesicle fusion specificity. Despite evidence suggestive of secondary losses amongst many lineages, the tethering complexes are well represented across the eukaryotes, suggesting an origin predating the radiation of eukaryotic lineages. Further, whilst we detect distant sequence relations between GARP, COG, exocyst and DSL1 components, these similarities most likely reflect convergent evolution of similar secondary structural elements. No similarity is found between the TRAPP and HOPS complexes and the other tethering factors. Overall, our data favour independent origins for the various tethering complexes. The taxa examined possess at least one homologue of each of the four SM protein families; since the four monophyletic families each encompass a wide diversity of eukaryotes, the SM protein families very likely evolved before the last common eukaryotic ancestor (LCEA).
Conclusion: These data further support a highly complex LCEA and indicate that the basic architecture of the trafficking system is remarkably conserved and ancient, with the SM proteins and tethering factors having originated very early in eukaryotic evolution. However, the independent origin of the tethering complexes suggests a novel pattern for increasing complexity in the membrane trafficking system, in addition to the pattern of paralogous machinery elaboration seen thus far.
Figures


References
-
- Stanier RY. The origins of photosynthesis in Eukaryotes. In: Carlile MJ and Skehel JJ, editor. Evolution in the Microbial World. Cambridge, U.K., Cambridge University Press; 1974. pp. 219–240.
-
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

