Virus membrane fusion
- PMID: 17320081
- PMCID: PMC7094569
- DOI: 10.1016/j.febslet.2007.01.093
Virus membrane fusion
Abstract
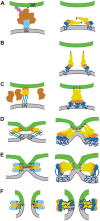
Membrane fusion of enveloped viruses with cellular membranes is mediated by viral glycoproteins (GP). Interaction of GP with cellular receptors alone or coupled to exposure to the acidic environment of endosomes induces extensive conformational changes in the fusion protein which pull two membranes into close enough proximity to trigger bilayer fusion. The refolding process provides the energy for fusion and repositions both membrane anchors, the transmembrane and the fusion peptide regions, at the same end of an elongated hairpin structure in all fusion protein structures known to date. The fusion process follows several lipidic intermediate states, which are generated by the refolding process. Although the major principles of viral fusion are understood, the structures of fusion protein intermediates and their mode of lipid bilayer interaction, the structures and functions of the membrane anchors and the number of fusion proteins required for fusion, necessitate further investigations.
Figures
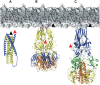
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

