Genetic characterization of 2,4,6-trichlorophenol degradation in Cupriavidus necator JMP134
- PMID: 17322325
- PMCID: PMC1892852
- DOI: 10.1128/AEM.02584-06
Genetic characterization of 2,4,6-trichlorophenol degradation in Cupriavidus necator JMP134
Abstract
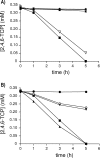
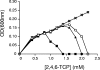
The degradation pathway of 2,4,6-trichlorophenol (2,4,6-TCP), a hazardous pollutant, in the aerobic bacterium Cupriavidus necator JMP134(pJP4) (formerly Ralstonia eutropha JMP134) is encoded by the tcp genes. These genes are located in a genetic context, tcpRXABCYD, which resembles a putative catabolic operon. In this work, these gene sequences were individually disrupted and mutant strains were evaluated for their ability to grow on or degrade 2,4,6-TCP. The tcpX and tcpA mutants completely failed to degrade this compound. Although the tcpC mutant was also unable to grow on 2,4,6-TCP, it still transformed this chlorophenol to 6-chlorohydroquinol. In contrast, the tcpD mutant grew on 2,4,6-TCP, suggesting the presence of additional maleylacetate reductase-encoding genes. Five other open reading frames encoding maleylacetate reductases, in addition to the tcpD gene, were found in the genome of C. necator, and two of them provide this function in the tcpD mutant. The tcpR gene, encoding a putative LysR-type transcriptional regulator, was disrupted, and this mutant strain completely failed to grow on 2,4,6-TCP. Transcriptional fusion studies demonstrated that TcpR activates the expression of the tcp genes, responding specifically to 2,4,6-TCP. The transcriptional start of the tcp operon was mapped, and a putative sigma(70)-type promoter was identified.
Figures


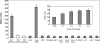
Similar articles
-
Efficient degradation of 2,4,6-Trichlorophenol requires a set of catabolic genes related to tcp genes from Ralstonia eutropha JMP134(pJP4).Appl Environ Microbiol. 2003 Dec;69(12):7108-15. doi: 10.1128/AEM.69.12.7108-7115.2003. Appl Environ Microbiol. 2003. PMID: 14660355 Free PMC article.
-
Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134.J Bacteriol. 2002 Jul;184(13):3492-500. doi: 10.1128/JB.184.13.3492-3500.2002. J Bacteriol. 2002. PMID: 12057943 Free PMC article.
-
Genuine genetic redundancy in maleylacetate-reductase-encoding genes involved in degradation of haloaromatic compounds by Cupriavidus necator JMP134.Microbiology (Reading). 2009 Nov;155(Pt 11):3641-3651. doi: 10.1099/mic.0.032086-0. Epub 2009 Aug 14. Microbiology (Reading). 2009. PMID: 19684066
-
Functions of flavin reductase and quinone reductase in 2,4,6-trichlorophenol degradation by Cupriavidus necator JMP134.J Bacteriol. 2008 Mar;190(5):1615-9. doi: 10.1128/JB.01697-07. Epub 2007 Dec 28. J Bacteriol. 2008. PMID: 18165297 Free PMC article.
-
Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134.FEMS Microbiol Rev. 2008 Aug;32(5):736-94. doi: 10.1111/j.1574-6976.2008.00122.x. Epub 2008 Aug 7. FEMS Microbiol Rev. 2008. PMID: 18691224 Review.
Cited by
-
Update of the risk assessment of brominated phenols and their derivatives in food.EFSA J. 2024 Oct 23;22(10):e9034. doi: 10.2903/j.efsa.2024.9034. eCollection 2024 Oct. EFSA J. 2024. PMID: 39444985 Free PMC article.
-
Structures of the inducer-binding domain of pentachlorophenol-degrading gene regulator PcpR from Sphingobium chlorophenolicum.Int J Mol Sci. 2014 Nov 12;15(11):20736-52. doi: 10.3390/ijms151120736. Int J Mol Sci. 2014. PMID: 25397598 Free PMC article.
-
Metabolism of 2-chloro-4-nitroaniline via novel aerobic degradation pathway by Rhodococcus sp. strain MB-P1.PLoS One. 2013 Apr 17;8(4):e62178. doi: 10.1371/journal.pone.0062178. Print 2013. PLoS One. 2013. Retraction in: PLoS One. 2014 Jul 09;9(7):e102862. doi: 10.1371/journal.pone.0102862. PMID: 23614030 Free PMC article. Retracted.
-
Novel gene clusters and metabolic pathway involved in 3,5,6-trichloro-2-pyridinol degradation by Ralstonia sp. strain T6.Appl Environ Microbiol. 2013 Dec;79(23):7445-53. doi: 10.1128/AEM.01817-13. Epub 2013 Sep 20. Appl Environ Microbiol. 2013. PMID: 24056464 Free PMC article.
-
Complete Genome Sequence of Ralstonia pickettii DTP0602, a 2,4,6-Trichlorophenol Degrader.Genome Announc. 2013 Oct 31;1(6):e00903-13. doi: 10.1128/genomeA.00903-13. Genome Announc. 2013. PMID: 24179121 Free PMC article.
References
-
- Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl (ed.). 1992. Short protocols in molecular biology, 2nd ed. Greene Publishing Associates, New York, NY.
-
- Briglia, M., F. A. Rainey, E. Stackebrandt, G. Schraa, and M. S. Salkinoja-Salonen. 1996. Rhodococcus percolatus sp. nov., a bacterium degrading 2,4,6-trichlorophenol. Int. J. Syst. Bacteriol. 46:23-30. - PubMed
-
- Clément, P., V. Matus, L. Cárdenas, and B. González. 1995. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol. Lett. 127:51-55. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

