Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium
- PMID: 17322329
- PMCID: PMC1855617
- DOI: 10.1128/AEM.00005-07
Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium
Abstract
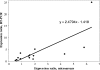
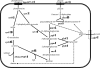
Lactobacillus helveticus CNRZ32 is used by the dairy industry to modulate cheese flavor. The compilation of a draft genome sequence for this strain allowed us to identify and completely sequence 168 genes potentially important for the growth of this organism in milk or for cheese flavor development. The primary aim of this study was to investigate the expression of these genes during growth in milk and MRS medium by using microarrays. Oligonucleotide probes against each of the completely sequenced genes were compiled on maskless photolithography-based DNA microarrays. Additionally, the entire draft genome sequence was used to produce tiled microarrays in which noninterrupted sequence contigs were covered by consecutive 24-mer probes and associated mismatch probe sets. Total RNA isolated from cells grown in skim milk or in MRS to mid-log phase was used as a template to synthesize cDNA, followed by Cy3 labeling and hybridization. An analysis of data from annotated gene probes identified 42 genes that were upregulated during the growth of CNRZ32 in milk (P < 0.05), and 25 of these genes showed upregulation after applying Bonferroni's adjustment. The tiled microarrays identified numerous additional genes that were upregulated in milk versus MRS. Collectively, array data showed the growth of CNRZ32 in milk-induced genes encoding cell-envelope proteinases, oligopeptide transporters, and endopeptidases as well as enzymes for lactose and cysteine pathways, de novo synthesis, and/or salvage pathways for purines and pyrimidines and other functions. Genes for a hypothetical phosphoserine utilization pathway were also differentially expressed. Preliminary experiments indicate that cheese-derived, phosphoserine-containing peptides increase growth rates of CNRZ32 in a chemically defined medium. These results suggest that phosphoserine is used as an energy source during the growth of L. helveticus CNRZ32.
Figures
References
-
- Bartels, H. J., M. E. Johnson, and N. F. Olson. 1987. Accelerated ripening of Gouda cheese. 2. Effect of freeze-shocked Lactobacillus helveticus on proteolysis and flavor development. Milchwissenschaft 42:139-144.
-
- Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. - PubMed
-
- Branny, P., F. de la Torre, and J. R. Garel. 1998. An operon encoding three glycolytic enzymes in Lactobacillus delbrueckii subsp. bulgaricus: glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase and triosephosphate isomerase. Microbiology 144:905-914. - PubMed
-
- Christensen, J. E., E. G. Dudley, J. A. Pederson, and J. L. Steele. 1999. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek 76:217-246. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

