Finely orchestrated movements: evolution of the ribosomal RNA genes
- PMID: 17322354
- PMCID: PMC1800602
- DOI: 10.1534/genetics.107.071399
Finely orchestrated movements: evolution of the ribosomal RNA genes
Abstract
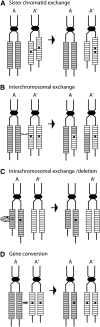
Evolution of the tandemly repeated ribosomal RNA (rRNA) genes is intriguing because in each species all units within the array are highly uniform in sequence but that sequence differs between species. In this review we summarize the origins of the current models to explain this process of concerted evolution, emphasizing early studies of recombination in yeast and more recent studies in Drosophila and mammalian systems. These studies suggest that unequal crossover is the major driving force in the evolution of the rRNA genes with sister chromatid exchange occurring more often than exchange between homologs. Gene conversion is also believed to play a role; however, direct evidence for its involvement has not been obtained. Remarkably, concerted evolution is so well orchestrated that even transposable elements that insert into a large fraction of the rRNA genes appear to have little effect on the process. Finally, we summarize data that suggest that recombination in the rDNA locus of higher eukaryotes is sufficiently frequent to monitor changes within a few generations.
Figures

Similar articles
-
Role of recombination in the long-term retention of transposable elements in rRNA gene loci.Genetics. 2008 Nov;180(3):1617-26. doi: 10.1534/genetics.108.093716. Epub 2008 Sep 14. Genetics. 2008. PMID: 18791229 Free PMC article.
-
Concerted evolution in the ribosomal RNA genes of an Epichloë endophyte hybrid: comparison between tandemly arranged rDNA and dispersed 5S rrn genes.Fungal Genet Biol. 2002 Feb;35(1):39-51. doi: 10.1006/fgbi.2001.1309. Fungal Genet Biol. 2002. PMID: 11860264
-
Gene conversion drives within genic sequences: concerted evolution of ribosomal RNA genes in bacteria and archaea.J Mol Evol. 2000 Oct;51(4):305-17. doi: 10.1007/s002390010093. J Mol Evol. 2000. PMID: 11040282
-
The role of selfish genetic elements in eukaryotic evolution.Nat Rev Genet. 2001 Aug;2(8):597-606. doi: 10.1038/35084545. Nat Rev Genet. 2001. PMID: 11483984 Review.
-
Evolution of R1 and R2 in the rDNA units of the genus Drosophila.Genetica. 1997;100(1-3):49-61. Genetica. 1997. PMID: 9440258 Review.
Cited by
-
Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA.PLoS One. 2012;7(8):e43971. doi: 10.1371/journal.pone.0043971. Epub 2012 Aug 30. PLoS One. 2012. PMID: 22952830 Free PMC article.
-
Analyses of chromosome copy number and expression level of four genes in the ciliate Chilodonella uncinata reveal a complex pattern that suggests epigenetic regulation.Gene. 2012 Aug 10;504(2):303-8. doi: 10.1016/j.gene.2012.04.067. Epub 2012 May 12. Gene. 2012. PMID: 22588027 Free PMC article.
-
Data mining reveals tissue-specific expression and host lineage-associated forms of Apis mellifera filamentous virus.PeerJ. 2023 Nov 14;11:e16455. doi: 10.7717/peerj.16455. eCollection 2023. PeerJ. 2023. PMID: 38025724 Free PMC article.
-
Non-allelic gene conversion enables rapid evolutionary change at multiple regulatory sites encoded by transposable elements.Elife. 2015 Feb 17;4:e05899. doi: 10.7554/eLife.05899. Elife. 2015. PMID: 25688566 Free PMC article.
-
Abundance, efficiency, and stability of reference transcript expression in a seasonal rodent: The Siberian hamster.PLoS One. 2022 Oct 3;17(10):e0275263. doi: 10.1371/journal.pone.0275263. eCollection 2022. PLoS One. 2022. PMID: 36190976 Free PMC article.
References
-
- Brown, D. D., P. C. Wensink and E. Jordan, 1972. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J. Mol. Biol. 63: 57–73. - PubMed
-
- Brutlag, D. L., 1980. Molecular arrangement and evolution of heterochromatic DNA. Annu. Rev. Genet. 14: 121–144. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

