Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo
- PMID: 17322397
- PMCID: PMC1804326
- DOI: 10.1101/gad.1509607
Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo
Abstract
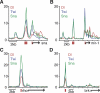
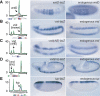
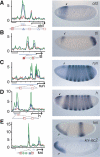
Genetic studies have identified numerous sequence-specific transcription factors that control development, yet little is known about their in vivo distribution across animal genomes. We determined the genome-wide occupancy of the dorsoventral (DV) determinants Dorsal, Twist, and Snail in the Drosophila embryo using chromatin immunoprecipitation coupled with microarray analysis (ChIP-chip). The in vivo binding of these proteins correlate tightly with the limits of known enhancers. Our analysis predicts substantially more target genes than previous estimates, and includes Dpp signaling components and anteroposterior (AP) segmentation determinants. Thus, the ChIP-chip data uncover a much larger than expected regulatory network, which integrates diverse patterning processes during development.
Figures


References
-
- Biemar F., Zinzen R., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Zinzen R., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Sementchenko V., Manak J.R., Levine M.S., Manak J.R., Levine M.S., Levine M.S. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc. Natl. Acad. Sci. 2005;102:15907–15911. - PMC - PubMed
-
- Biemar F., Nix D.A., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Nix D.A., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Ronshaugen M., Sementchenko V., Bell I., Manak J.R., Levine M.S., Sementchenko V., Bell I., Manak J.R., Levine M.S., Bell I., Manak J.R., Levine M.S., Manak J.R., Levine M.S., Levine M.S. Comprehensive identification of Drosophila dorsal–ventral patterning genes using a whole-genome tiling array. Proc. Natl. Acad. Sci. 2006;103:12763–12768. - PMC - PubMed
-
- Butler B.A., Soong J., Gergen J.P., Soong J., Gergen J.P., Gergen J.P. The Drosophila segmentation gene runt has an extended cis-regulatory region that is required for vital expression at other stages of development. Mech. Dev. 1992;39:17–28. - PubMed
-
- Carroll S.B., Winslow G.M., Twombly V.J., Scott M.P., Winslow G.M., Twombly V.J., Scott M.P., Twombly V.J., Scott M.P., Scott M.P. Genes that control dorsoventral polarity affect gene expression along the anteroposterior axis of the Drosophilaembryo. Development. 1987;99:327–332. - PubMed
-
- Chanas G., Lavrov S., Iral F., Cavalli G., Maschat F., Lavrov S., Iral F., Cavalli G., Maschat F., Iral F., Cavalli G., Maschat F., Cavalli G., Maschat F., Maschat F. Engrailed and polyhomeotic maintain posterior cell identity through cubitus-interruptus regulation. Dev. Biol. 2004;272:522–535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
