Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis
- PMID: 17322408
- PMCID: PMC1867327
- DOI: 10.1105/tpc.106.047563
Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis
Abstract
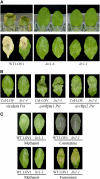
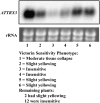
The fungus Cochliobolus victoriae causes Victoria blight of oats (Avena sativa) and is pathogenic due to its production of victorin, which induces programmed cell death in sensitive plants. Victorin sensitivity has been identified in Arabidopsis thaliana and is conferred by the dominant gene LOCUS ORCHESTRATING VICTORIN EFFECTS1 (LOV1), which encodes a coiled-coil-nucleotide binding site-leucine-rich repeat protein. We isolated 63 victorin-insensitive mutants, including 59 lov1 mutants and four locus of insensitivity to victorin1 (liv1) mutants. The LIV1 gene encodes thioredoxin h5 (ATTRX5), a member of a large family of disulfide oxidoreductases. To date, very few plant thioredoxins have been assigned specific, nonredundant functions. We found that the victorin response was highly specific to ATTRX5, as the closely related ATTRX3 could only partially compensate for loss of ATTRX5, even when overexpressed. We also created chimeric ATTRX5/ATTRX3 proteins, which identified the central portion of the protein as important for conferring specificity to ATTRX5. Furthermore, we found that ATTRX5, but not ATTRX3, is highly induced in sensitive Arabidopsis following victorin treatment. Finally, we determined that only the first of the two active-site Cys residues in ATTRX5 is required for the response to victorin, suggesting that ATTRX5 function in the victorin pathway involves an atypical mechanism of action.
Figures




Similar articles
-
Characterization of natural and induced variation in the LOV1 gene, a CC-NB-LRR gene conferring victorin sensitivity and disease susceptibility in Arabidopsis.Mol Plant Microbe Interact. 2008 Jan;21(1):7-19. doi: 10.1094/MPMI-21-1-0007. Mol Plant Microbe Interact. 2008. PMID: 18052878
-
Characterization of the LOV1-mediated, victorin-induced, cell-death response with virus-induced gene silencing.Mol Plant Microbe Interact. 2013 Aug;26(8):903-17. doi: 10.1094/MPMI-01-13-0014-R. Mol Plant Microbe Interact. 2013. PMID: 23634836
-
Identification and characterization of victorin sensitivity in Arabidopsis thaliana.Mol Plant Microbe Interact. 2004 Jun;17(6):577-82. doi: 10.1094/MPMI.2004.17.6.577. Mol Plant Microbe Interact. 2004. PMID: 15195940
-
The thioredoxin h system of higher plants.Plant Physiol Biochem. 2004 Apr;42(4):265-71. doi: 10.1016/j.plaphy.2004.03.002. Plant Physiol Biochem. 2004. PMID: 15120110 Review.
-
The plant thioredoxin system.Cell Mol Life Sci. 2005 Jan;62(1):24-35. doi: 10.1007/s00018-004-4296-4. Cell Mol Life Sci. 2005. PMID: 15619004 Free PMC article. Review.
Cited by
-
Identification of candidate genes responsible for the susceptibility of apple (Malus × domestica Borkh.) to Alternaria blotch.BMC Plant Biol. 2019 Apr 8;19(1):132. doi: 10.1186/s12870-019-1737-7. BMC Plant Biol. 2019. PMID: 30961541 Free PMC article.
-
Synthesis of redox-active molecules and their signaling functions during the expression of plant disease resistance.Antioxid Redox Signal. 2013 Sep 20;19(9):990-7. doi: 10.1089/ars.2013.5429. Epub 2013 Jul 17. Antioxid Redox Signal. 2013. PMID: 23725342 Free PMC article. Review.
-
The Ralstonia solanacearum type III effector RipAY targets plant redox regulators to suppress immune responses.Mol Plant Pathol. 2018 Jan;19(1):129-142. doi: 10.1111/mpp.12504. Epub 2016 Dec 27. Mol Plant Pathol. 2018. PMID: 27768829 Free PMC article.
-
Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia.Plant Physiol. 2009 Aug;150(4):1733-49. doi: 10.1104/pp.109.140251. Epub 2009 Jul 1. Plant Physiol. 2009. PMID: 19571308 Free PMC article.
-
Functional Characterization of Lobularia maritima LmTrxh2 Gene Involved in Cold Tolerance in Tobacco through Alleviation of ROS Damage to the Plasma Membrane.Int J Mol Sci. 2023 Feb 3;24(3):3030. doi: 10.3390/ijms24033030. Int J Mol Sci. 2023. PMID: 36769352 Free PMC article.
References
-
- Balmer, Y., Vensel, W.H., Tanaka, C.K., Hurkman, W.J., Gelhaye, E., Rouhier, N., Jacquot, J.-P., Manieri, W., Schürmann, P., Droux, M., and Buchanan, B.B. (2004). Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. USA 101 2642–2647. - PMC - PubMed
-
- Belkhadir, Y., Subramaniam, R., and Dangl, J.L. (2004). Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7 391–399. - PubMed
-
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265 1856–1860. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

