In vitro ligation of oligodeoxynucleotides containing C8-oxidized purine lesions using bacteriophage T4 DNA ligase
- PMID: 17323928
- PMCID: PMC2442820
- DOI: 10.1021/bi062214k
In vitro ligation of oligodeoxynucleotides containing C8-oxidized purine lesions using bacteriophage T4 DNA ligase
Abstract
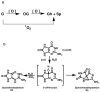
Ligases conduct the final stage of repair of DNA damage by sealing a single-stranded nick after excision of damaged nucleotides and reinsertion of correct nucleotides. Depending upon the circumstances and the success of the repair process, lesions may remain at the ligation site, either in the template or at the oligomer termini to be joined. Ligation experiments using bacteriophage T4 DNA ligase were carried out with purine lesions in four positions surrounding the nick site in a total of 96 different duplexes. The oxidized lesion 8-oxo-7,8-dihydroguanosine (OG) showed, as expected, that the enzyme is most sensitive to lesions on the 3' end of the nick compared to the 5' end and to lesions located in the intact template strand. In general, substrates containing the OG.A mismatch were more readily ligated than those with the OG.C mismatch. Ligations of duplexes containing the OA.T base pair (OA = 8-oxo-7,8-dihydroadenosine) that could adopt an anti-anti conformation proceeded with high efficiencies. An OI.A mismatch-containing duplex (OI = 8-oxo-7,8-dihydroinosine) behaved like OG.A. Due to its low reduction potential, OG is readily oxidized to secondary oxidation products, such as the guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) nucleosides; these lesions also contain an oxo group at the original C8 position of the purine. Ligation of oligomers containing Gh and Sp occurred when opposite A and G, although the overall ligation efficiencies were much lower than those of most OG base pairs. Steady-state kinetic studies were carried out for representative examples of lesions in the template. Km increased by 90-100-fold for OG.C-, OI.C-, OI.A-, and OA.T-containing duplexes compared to that of a G.C-containing duplex. Substrates containing Gh.A, Gh.G, Sp.A, and Sp.G base pairs exhibited Km values 20-70-fold higher than that of the substrate containing a G.C base pair, while the Km value for OG.A was 5 times lower than that for G.C.
Figures
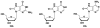


Similar articles
-
Recognition and removal of oxidized guanines in duplex DNA by the base excision repair enzymes hOGG1, yOGG1, and yOGG2.Biochemistry. 2003 Sep 30;42(38):11373-81. doi: 10.1021/bi034951b. Biochemistry. 2003. PMID: 14503888
-
Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG.Biochemistry. 2000 Dec 5;39(48):14984-92. doi: 10.1021/bi0017982. Biochemistry. 2000. PMID: 11101315
-
In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment).Biochemistry. 2002 Dec 24;41(51):15304-14. doi: 10.1021/bi0264925. Biochemistry. 2002. PMID: 12484769
-
Structure and potential mutagenicity of new hydantoin products from guanosine and 8-oxo-7,8-dihydroguanine oxidation by transition metals.Environ Health Perspect. 2002 Oct;110 Suppl 5(Suppl 5):713-7. doi: 10.1289/ehp.02110s5713. Environ Health Perspect. 2002. PMID: 12426118 Free PMC article. Review.
-
Formation and processing of DNA damage substrates for the hNEIL enzymes.Free Radic Biol Med. 2017 Jun;107:35-52. doi: 10.1016/j.freeradbiomed.2016.11.030. Epub 2016 Nov 20. Free Radic Biol Med. 2017. PMID: 27880870 Free PMC article. Review.
Cited by
-
Reverse Transcription Past Products of Guanine Oxidation in RNA Leads to Insertion of A and C opposite 8-Oxo-7,8-dihydroguanine and A and G opposite 5-Guanidinohydantoin and Spiroiminodihydantoin Diastereomers.Biochemistry. 2017 Sep 26;56(38):5053-5064. doi: 10.1021/acs.biochem.7b00730. Epub 2017 Sep 11. Biochemistry. 2017. PMID: 28845978 Free PMC article.
-
Effects of 3'-OH and 5'-PO4 base mispairs and damaged base lesions on the fidelity of nick sealing by Deinococcus radiodurans RNA ligase.J Bacteriol. 2014 May;196(9):1704-12. doi: 10.1128/JB.00020-14. Epub 2014 Feb 14. J Bacteriol. 2014. PMID: 24532777 Free PMC article.
-
Unusual structural features of hydantoin lesions translate into efficient recognition by Escherichia coli Fpg.Biochemistry. 2007 Aug 21;46(33):9355-65. doi: 10.1021/bi602459v. Epub 2007 Jul 27. Biochemistry. 2007. PMID: 17655276 Free PMC article.
-
Kinetic mechanism and fidelity of nick sealing by Escherichia coli NAD+-dependent DNA ligase (LigA).Nucleic Acids Res. 2016 Mar 18;44(5):2298-309. doi: 10.1093/nar/gkw049. Epub 2016 Feb 8. Nucleic Acids Res. 2016. PMID: 26857547 Free PMC article.
-
Inhibition of DNA glycosylases via small molecule purine analogs.PLoS One. 2013 Dec 9;8(12):e81667. doi: 10.1371/journal.pone.0081667. eCollection 2013. PLoS One. 2013. PMID: 24349107 Free PMC article.
References
-
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. - PubMed
-
- Feig DI, Loeb LA. Oxygen radical induced mutagenesis is DNA polymerase specific. J Mol Biol. 1994;235:33–41. - PubMed
-
- Steenken S, Jovanovic SJ. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

